Section 4 Point Cloud Processing and Pile Segmentation
To start, we’ll use the point cloud data alone to attempt to classify slash piles using structural signatures alone without additional spectral information.
We’ll use the cloud2trees package to perform all preprocessing of point cloud data which includes:
- ground classification and noise removal
- raster data (DTM and CHM) generation
- point cloud height normalization
All of this can be accomplished using the cloud2raster() function. After generating these products from the raw point cloud we’ll perform object segmentation to attempt to detect round, conical objects like slash piles from: 1) the normalized point cloud directly; 2) the CHM which we’ll generate by setting the minimum height to zero (essentially a digital surface model [DSM] with the ground removed).
To attempt to identify slash piles from the CHM/DSM, we can use watershed segmentation in a bottom-up approach. Insert something from paper about bottom-up approach that uses a CHM “slice” near the ground. For slash piles, which are often irregular and may not have a distinct “treetop” equivalent, CHM-based methods might be less directly applicable unless the piles present a very clear, isolated conical or rounded form. Could use expected morphology of the slash piles (e.g. maximum height) based on prior research and/or the pile construction prescription. We won’t use the height normalized point cloud data in this project but future work could attempt to detect slash piles directly from the point cloud by using a clustering algorithm such as DBSCAN (Density-Based Spatial Clustering of Applications with Noise), for example.
- Section 5 builds and tests a raster-based watershed segmentation methodology
- Section 6 combines the raster-based watershed segmentation methodology with spectral information in a data fusion approach
4.1 Process Raw Point Cloud
We’ll use cloud2trees::cloud2raster() to process the raw point cloud data
# set chm res
chm_res_m_temp <- 0.2
dir_temp <- paste0("../data/point_cloud_processing_delivery_chm",chm_res_m_temp,"m")
# do it
if(!dir.exists(dir_temp)){
# cloud2trees
cloud2raster_ans <- cloud2trees::cloud2raster(
output_dir = "../data"
, input_las_dir = "f:\\PFDP_Data\\p4pro_images\\P4Pro_06_17_2021_half_half_optimal\\2_densification\\point_cloud"
, accuracy_level = 2
, keep_intrmdt = T
, dtm_res_m = 0.25
, chm_res_m = chm_res_m_temp
, min_height = 0 # effectively generates a DSM based on non-ground points
)
# rename
file.rename(from = "../data/point_cloud_processing_delivery", to = dir_temp)
}else{
dtm_temp <- terra::rast( file.path(dir_temp, "dtm_0.25m.tif") )
chm_temp <- terra::rast( file.path(dir_temp, paste0("chm_", chm_res_m_temp,"m.tif")) )
# dtm_temp <- terra::rast("../data/point_cloud_processing_delivery_chm0.1m/dtm_0.25m.tif")
# chm_temp <- terra::rast("../data/point_cloud_processing_delivery_chm0.1m/chm_0.1m.tif")
# dtm_temp <- terra::rast("../data/point_cloud_processing_delivery_chm0.2m/dtm_0.5m.tif")
# chm_temp <- terra::rast("../data/point_cloud_processing_delivery_chm0.2m/chm_0.2m.tif")
cloud2raster_ans <- list(
"dtm_rast" = dtm_temp
, "chm_rast" = chm_temp
)
}let’s check out the DTM
cloud2raster_ans$dtm_rast %>%
terra::aggregate(fact = 2) %>% #, fun = "median", cores = lasR::half_cores(), na.rm = T) %>%
terra::plot(axes = F)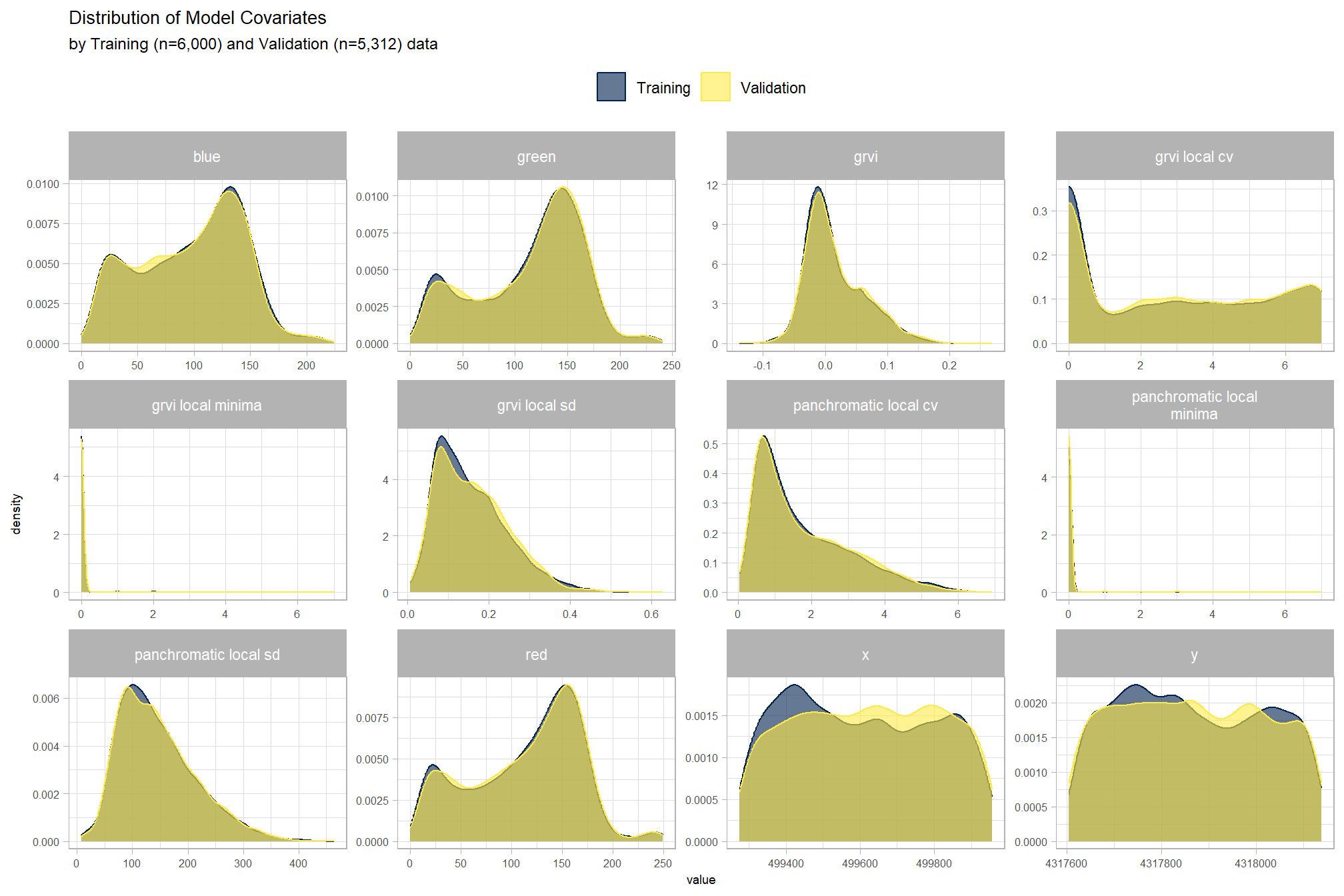
and CHM
cloud2raster_ans$chm_rast %>%
terra::aggregate(fact = 2) %>% #, fun = "median", cores = lasR::half_cores(), na.rm = T) %>%
terra::plot(col = viridis::plasma(100), axes = F)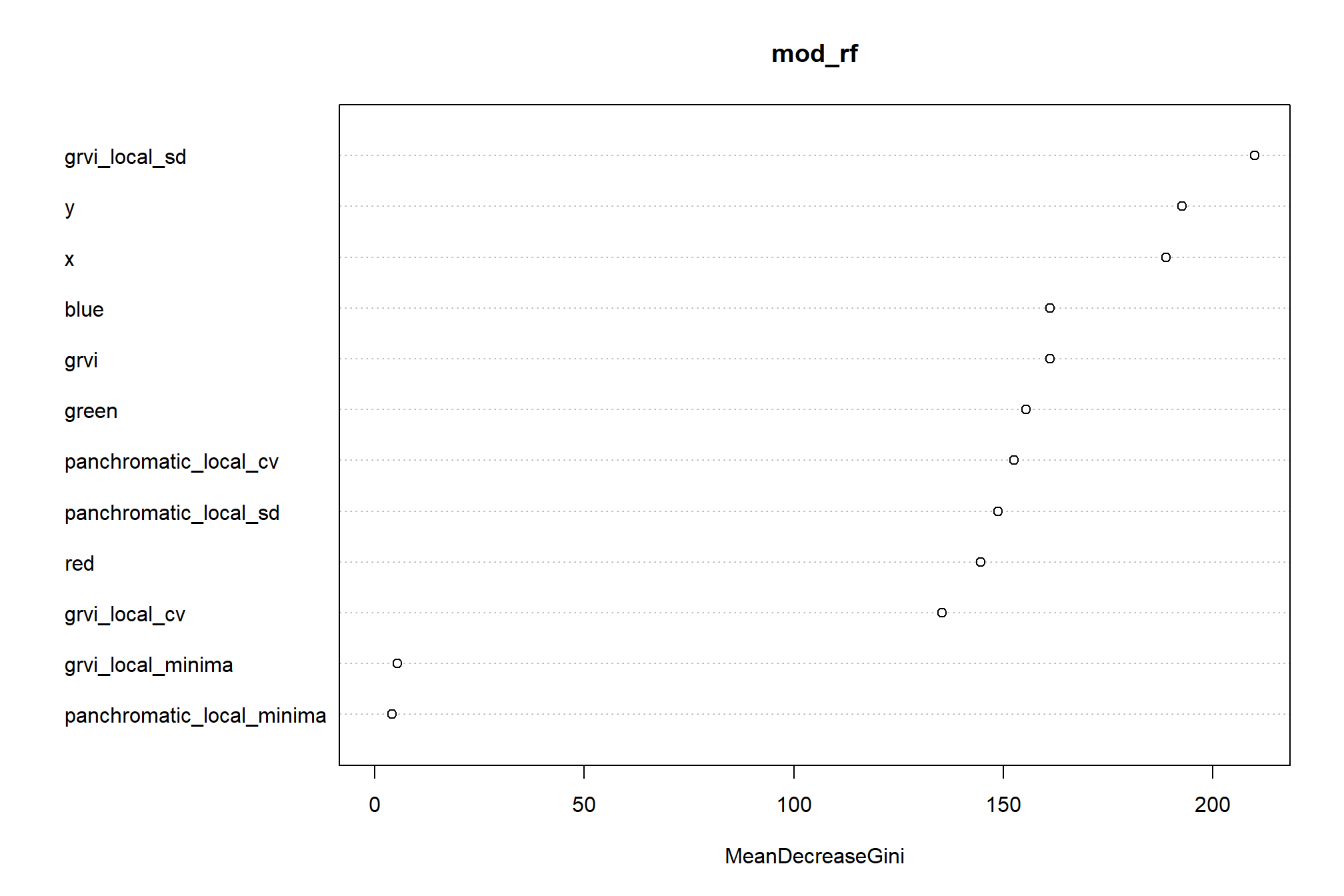
4.1.1 DTM and CHM + piles
let’s visually inspect the DTM and CHM with the pile outlines overlaid
p_fn_temp <- function(
rn
, df = slash_piles_polys
, rast = cloud2raster_ans$dtm_rast
, crs = terra::crs(cloud2raster_ans$dtm_rast)
, my_title = ""
, vopt = "viridis"
, lim = NULL
) {
# scale the buffer based on the largest
d_temp <- df %>%
dplyr::arrange(tolower(comment), desc(field_diameter_m)) %>%
dplyr::slice(rn) %>%
sf::st_transform(crs)
# plt classifier
# convert to stars
comp_st <- rast %>%
terra::crop(
d_temp %>% sf::st_buffer(20) %>% terra::vect()
) %>%
terra::as.data.frame(xy = T) %>%
dplyr::rename(f=3)
# ggplot
comp_temp <- ggplot2::ggplot() +
ggplot2::geom_tile(data = comp_st, ggplot2::aes(x=x,y=y,fill=f)) +
ggplot2::geom_sf(data = d_temp, fill = NA, color = "gray33", lwd = 0.4) +
ggplot2::scale_x_continuous(expand = c(0, 0)) +
ggplot2::scale_y_continuous(expand = c(0, 0)) +
ggplot2::labs(
x = ""
, y = ""
, subtitle = my_title
) +
ggplot2::theme_void() +
ggplot2::theme(
legend.position = "none" # c(0.5,1)
, legend.margin = margin(0,0,0,0)
, legend.text = element_text(size = 8)
, legend.title = element_text(size = 8)
, legend.key = element_rect(fill = "white")
# , plot.title = ggtext::element_markdown(size = 10, hjust = 0.5)
, plot.title = element_text(size = 8, hjust = 0.5, face = "bold")
, plot.subtitle = element_text(size = 6, hjust = 0.5, face = "italic")
)
if(!is.null(lim)){
comp_temp <- comp_temp +
ggplot2::scale_fill_viridis_c(option = vopt, limits = lim)
}else{
comp_temp <- comp_temp +
ggplot2::scale_fill_viridis_c(option = vopt)
}
plt_temp <- comp_temp
return(list("plt"=plt_temp,"d"=d_temp))
}
# p_fn_temp(
# rn = 5
# # , lim = c(floor(terra::minmax(cloud2raster_ans$dtm_rast)[1]*0.98), floor(terra::minmax(cloud2raster_ans$dtm_rast)[2]*1.02))
# )
# p_fn_temp(
# rn = 11
# , rast = cloud2raster_ans$chm_rast
# , vopt = "plasma"
# )
# combine 3
plt_combine_temp <- function(
rn
, df = slash_piles_polys %>% dplyr::filter(!is.na(comment))
, rast1 = cloud2raster_ans$dtm_rast
, title1 = "DTM"
, vopt1 = "viridis"
, rast2 = cloud2raster_ans$chm_rast
, title2 = "CHM"
, vopt2 = "plasma"
, crs = terra::crs(cloud2raster_ans$dtm_rast)
) {
# composite 1
ans1 <- p_fn_temp(
rn = rn
, df = df
, rast = rast1
, my_title = title1
, crs = crs
, vopt = vopt1
)
# composite 2
ans2 <- p_fn_temp(
rn = rn
, df = df
, rast = rast2
, my_title = title2
, crs = crs
, vopt = vopt2
)
# plt rgb
rgb_temp <-
ortho_plt_fn(my_ortho_rast = ortho_rast, stand =ans1$d) +
ggplot2::geom_sf(data = ans1$d, fill = NA, color = "white", lwd = 0.3)
# combine
r <- ans1$plt + ans2$plt + rgb_temp
return(r)
}
# plt_combine_temp(2)
# add pile locations
plt_list_rast_comp <- sample(1:nrow(slash_piles_polys %>% dplyr::filter(!is.na(comment))),10) %>%
purrr::map(plt_combine_temp)
# plt_list_rast_comp[[2]]combine the plots for a few piles
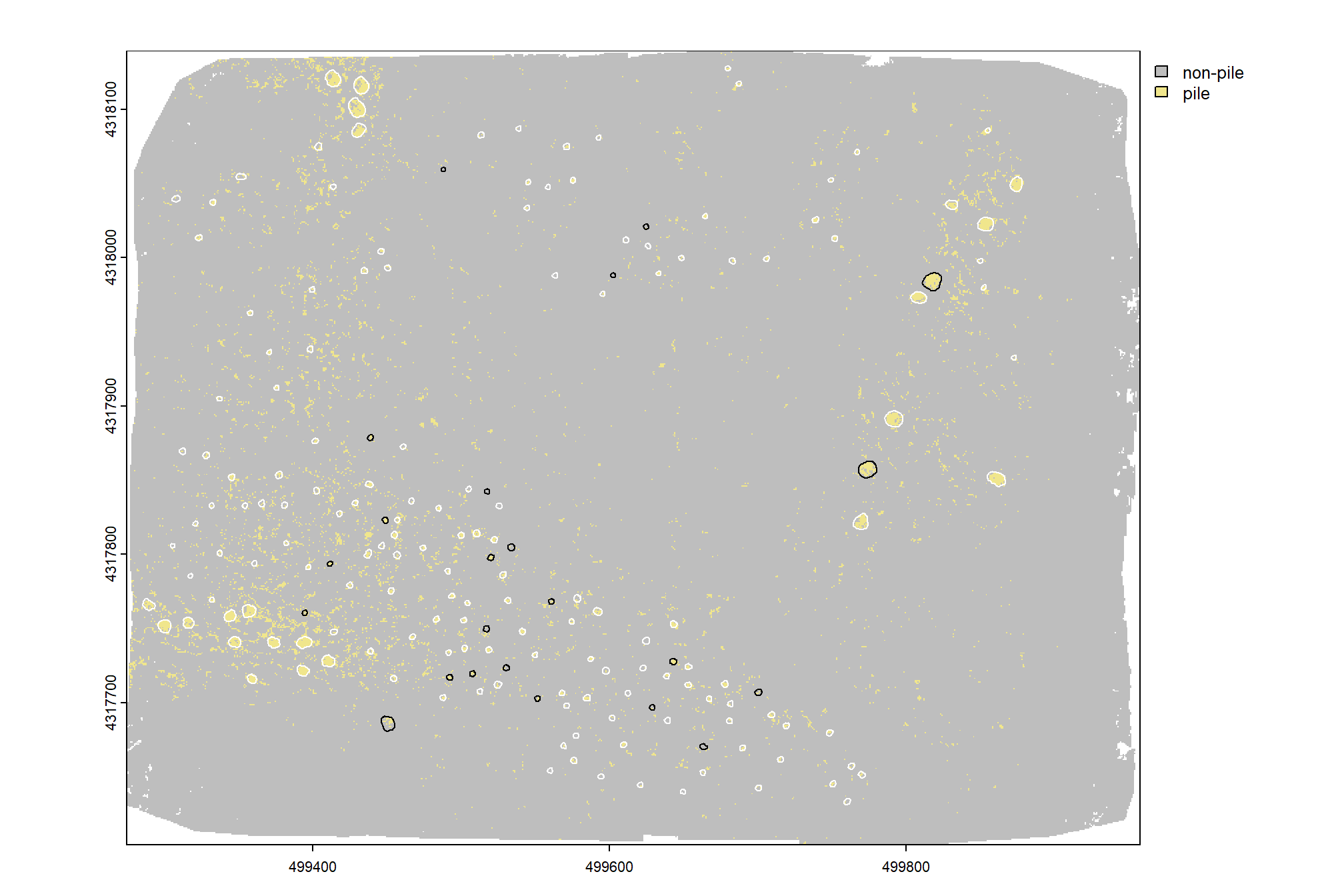
a few things are noteworthy in these examples:
- Piles are clearly visible in some areas of the DTM but less visible in other areas
- Piles are delineated in the CHM but with varying degrees of definition based on surrounding terrain and pile structure
- The DTM and CHM were rarely impacted by shadows in the RGB imagery
- It is interesting to see coarse woody debris occasionally visible in the CHM