Chapter 4 Multilevel linear models: varying slopes, non-nested models, and other complexities (Ch 13)
This chapter starts on page 279
4.1 Setup
# bread-and-butter
library(tidyverse)
library(lubridate)
library(viridis)
library(scales)
library(latex2exp)
# visualization
library(cowplot)
library(kableExtra)
# Linear Mixed-Effects Models
library(lme4)
library(broom.mixed)
# jags and bayesian
library(rjags)
library(MCMCvis)
library(HDInterval)
library(monomvn)
#set seed
set.seed(11)4.2 Load data
# load individual, house-level observations
# srrs2 <- read.table("http://www.stat.columbia.edu/~gelman/arm/examples/radon/srrs2.dat", header=TRUE, sep=",") %>%
srrs2 <- read.table("../data/radon/srrs2.dat", header=TRUE, sep=",") %>%
dplyr::mutate(
fullfips = stfips*1000 + cntyfips
)
# load county data
# cty <- read.table("http://www.stat.columbia.edu/~gelman/arm/examples/radon/cty.dat", header=TRUE, sep=",") %>%
cty <- read.table("../data/radon/cty.dat", header=TRUE, sep=",") %>%
dplyr::mutate(fullfips = stfips*1000 + ctfips) %>%
# there are duplicates in this data
dplyr::group_by(fullfips) %>%
dplyr::filter(dplyr::row_number()==1) %>%
dplyr::ungroup() %>%
dplyr::mutate(u = log(Uppm))
# join to individual, house radon measurement data
srrs2 <- srrs2 %>%
dplyr::left_join(cty %>% dplyr::select(fullfips, u), by = c("fullfips"="fullfips"))
# filter MN and create vars
radon_mn <- srrs2 %>%
dplyr::filter(
state=="MN"
) %>%
dplyr::mutate(
radon = activity
, log.radon = log(ifelse(radon==0, .1, radon))
, y = log.radon
, x = floor # 0 for basement, 1 for first floor
, county_index = as.numeric(as.factor(county))
)
# unique uranium by county index 1:85
u <- radon_mn %>%
dplyr::group_by(county_index) %>%
summarize(u = dplyr::first(u)) %>%
ungroup() %>%
dplyr::arrange(county_index)4.3 Varying intercepts and slopes (p. 279)
The next step in multilevel modeling is to allow more than one regression coefficient to vary by group. We shall illustrate with the radon model from the previous chapter, which is relatively simple because it only has a single individual-level predictor, \(x\) (the indicator for whether the measurement was taken on the first floor). We begin with a varying-intercept, varying-slope model including \(x\) but without the county-level uranium predictor; thus,
\[\begin{align*} y_i &\sim \textrm{N}(\alpha_{j[i]} + \beta_{j[i]} \cdot x_i, \sigma^2_y) \\ &\textrm{for} \; i = 1, \cdots, n \\ \left(\begin{array}{c} \alpha_{j}\\ \beta_{j} \end{array}\right) &\sim \textrm{N}\left(\left(\begin{array}{c} \mu_\alpha\\ \mu_\beta \end{array}\right) , \left(\begin{array}{cc} \sigma^{2}_{\alpha} & \rho\sigma_{\alpha}\sigma_{\beta}\\ \rho\sigma_{\alpha}\sigma_{\beta} & \sigma^{2}_{\beta} \end{array}\right) \right) \\ &\textrm{for} \; j = 1, \cdots, J \end{align*}\]
with variation in the \(\alpha_{j}\)’s and the \(\beta_{j}\)’s and also a between-group correlation parameter \(\rho\)
4.3.1 Using lme4::lmer (p. 279)
## Varying intercept & slopes w/ no group level predictors
M3 <- lme4::lmer(data = radon_mn, formula = y ~ x + (1 + x | county))
summary(M3)Linear mixed model fit by REML [‘lmerMod’] Formula: y ~ x + (1 + x | county) Data: radon_mn
REML criterion at convergence: 2168.3
Scaled residuals: Min 1Q Median 3Q Max -4.4044 -0.6224 0.0138 0.6123 3.5682
Random effects:
Groups Name Variance Std.Dev. Corr
county (Intercept) 0.1216 0.3487
x 0.1181 0.3436 -0.34
Residual 0.5567 0.7462
Number of obs: 919, groups: county, 85
Fixed effects: Estimate Std. Error t value (Intercept) 1.46277 0.05387 27.155 x -0.68110 0.08758 -7.777
Correlation of Fixed Effects: (Intr) x -0.381
#estimated regression coefficicents
coef(M3)$county[1:3,] (Intercept) xAITKIN 1.1445374 -0.5406207 ANOKA 0.9333795 -0.7708213 BECKER 1.4716909 -0.6688831
# fixed and random effects
fixef(M3)(Intercept) x 1.4627700 -0.6810982
ranef(M3)$county[1:3,] (Intercept) xAITKIN -0.318232574 0.14047747 ANOKA -0.529390508 -0.08972314 BECKER 0.008920884 0.01221507
# check intercept match for county 1
fixef(M3)["(Intercept)"] + ranef(M3)$county[1,"(Intercept)"](Intercept) 1.144537
coef(M3)$county[1,"(Intercept)"][1] 1.144537
# check slope match for county 1
fixef(M3)["x"] + ranef(M3)$county[1,"x"] x -0.5406207
coef(M3)$county[1,"x"][1] -0.5406207
4.3.2 Bayesian
4.3.2.1 Data and Initial conditions
# varying-intercept model, no predictors
## data for jags
data <- list(
n = nrow(radon_mn)
, J = radon_mn$county %>% unique() %>% length()
, y = radon_mn$log.radon %>% as.double()
, x = radon_mn$x %>% as.double()
, county = radon_mn$county_index %>% as.double()
)
## inits for jags
inits <- function(){
list(
B.matrix = array(data = rnorm(n=2*data$J, mean = 0, sd = 1), dim = c(data$J,2))
, mu.alpha = rnorm(1, mean = 0, sd = 1)
, mu.beta = rnorm(1, mean = 0, sd = 1)
, sigma.alpha = runif(n = 1, min = 0, max = 1)
, sigma.beta = runif(n = 1, min = 0, max = 1)
, sigma.y = runif(n = 1, min = 0, max = 1)
, rho = runif(n = 1, min = -1, max = 1)
)
}
# define parameters to return from MCMC
params <- c (
"alpha"
, "beta"
, "mu.alpha"
, "mu.beta"
, "sigma.alpha"
, "sigma.beta"
, "sigma.y"
, "rho"
)4.3.2.2 JAGS Model
Recall model defined above
Write out the JAGS code for the model.
## JAGS Model
model {
###########################
# priors
###########################
# coefficient of correlation for covariance matrix
rho ~ dunif(-1,1)
# alpha priors
mu.alpha ~ dnorm(0,1E-6)
sigma.alpha ~ dunif(0, 100)
tau.alpha <- 1/sigma.alpha^2
# beta priors
mu.beta ~ dnorm(0,1E-6)
sigma.beta ~ dunif(0, 100)
tau.beta <- 1/sigma.beta^2
# y priors
sigma.y ~ dunif(0, 100)
tau.y <- 1/sigma.y^2
###########################
# variance-covariance matrix
###########################
# variance
Sigma.matrix[1, 1] <- sigma.alpha^2 # row 1, column 1
Sigma.matrix[2, 2] <- sigma.beta^2 # row 2, column 2
# Cov(alpha,beta) = rho * sigma.alpha * sigma.beta
Sigma.matrix[2, 1] <- rho * sigma.alpha * sigma.beta # row 2, column 1
Sigma.matrix[1, 2] <- Sigma.matrix[2, 1] # row 1, column 2
# invert Sigma matrix
Omega.matrix <- inverse(Sigma.matrix)
###########################
# likelihood
###########################
# varying slopes and intercepts at the county level
for(j in 1:J){
# put mu.alpha and mu.beta in matrix
B.hat[j,1] <- mu.alpha # column 1
B.hat[j,2] <- mu.beta # column 2
# note multivariate normal
B.matrix[j,1:2] ~ dmnorm(B.hat[j,], Omega.matrix)
# extract alpha and beta from matrix
alpha[j] <- B.matrix[j, 1] # column 1
beta[j] <- B.matrix[j, 2] # column 2
}
# y
for (i in 1:n){
mu.y[i] <- alpha[county[i]] + beta[county[i]] * x[i]
y[i] ~ dnorm(mu.y[i], tau.y)
}
}4.3.2.3 Implement JAGS Model
##################################################################
# insert JAGS model code into an R script
##################################################################
{ # Extra bracket needed only for R markdown files - see answers
sink("intslp_pred.R") # This is the file name for the jags code
cat("
## JAGS Model
model {
###########################
# priors
###########################
# coefficient of correlation for covariance matrix
rho ~ dunif(-1,1)
# alpha priors
mu.alpha ~ dnorm(0,1E-6)
sigma.alpha ~ dunif(0, 100)
tau.alpha <- 1/sigma.alpha^2
# beta priors
mu.beta ~ dnorm(0,1E-6)
sigma.beta ~ dunif(0, 100)
tau.beta <- 1/sigma.beta^2
# y priors
sigma.y ~ dunif(0, 100)
tau.y <- 1/sigma.y^2
###########################
# variance-covariance matrix
###########################
# variance
Sigma.matrix[1, 1] <- sigma.alpha^2 # row 1, column 1
Sigma.matrix[2, 2] <- sigma.beta^2 # row 2, column 2
# Cov(alpha,beta) = rho * sigma.alpha * sigma.beta
Sigma.matrix[2, 1] <- rho * sigma.alpha * sigma.beta # row 2, column 1
Sigma.matrix[1, 2] <- Sigma.matrix[2, 1] # row 1, column 2
# invert Sigma matrix
Omega.matrix <- inverse(Sigma.matrix)
###########################
# likelihood
###########################
# varying slopes and intercepts at the county level
for(j in 1:J){
# put mu.alpha and mu.beta in matrix
B.hat[j,1] <- mu.alpha # column 1
B.hat[j,2] <- mu.beta # column 2
# note multivariate normal
B.matrix[j,1:2] ~ dmnorm(B.hat[j,], Omega.matrix)
# extract alpha and beta from matrix
alpha[j] <- B.matrix[j, 1] # column 1
beta[j] <- B.matrix[j, 2] # column 2
}
# y
for (i in 1:n){
mu.y[i] <- alpha[county[i]] + beta[county[i]] * x[i]
y[i] ~ dnorm(mu.y[i], tau.y)
}
}
", fill = TRUE)
sink()
}
##################################################################
# implement model
##################################################################
######################
# Call to JAGS
######################
intslp_pred = rjags::jags.model(
file = "intslp_pred.R"
, data = data
, inits = inits
, n.chains = 3
, n.adapt = 500
)## Compiling model graph
## Resolving undeclared variables
## Allocating nodes
## Graph information:
## Observed stochastic nodes: 919
## Unobserved stochastic nodes: 91
## Total graph size: 3410
##
## Initializing modelstats::update(intslp_pred, n.iter = 1000, progress.bar = "none")
# save the coda object (more precisely, an mcmc.list object) to R
mlm_intslp_pred = rjags::coda.samples(
model = intslp_pred
, variable.names = params
, n.iter = 2500
, n.thin = 1
, progress.bar = "none"
)4.3.2.4 Summary of the marginal posterior distributions of the parameters
# summary
MCMCvis::MCMCsummary(mlm_intslp_pred, params = params[!params %in% c("alpha", "beta")]) %>%
kableExtra::kable(
caption = "Bayesian: Varying-intercept & slope multilevel model, floor of measurement predictor"
, digits = 4
) %>%
kableExtra::kable_styling(font_size = 12)| mean | sd | 2.5% | 50% | 97.5% | Rhat | n.eff | |
|---|---|---|---|---|---|---|---|
| mu.alpha | 1.4614 | 0.0557 | 1.3541 | 1.4608 | 1.5701 | 1.01 | 1070 |
| mu.beta | -0.6812 | 0.0936 | -0.8475 | -0.6819 | -0.4939 | 1.02 | 286 |
| sigma.alpha | 0.3482 | 0.0493 | 0.2562 | 0.3468 | 0.4500 | 1.01 | 703 |
| sigma.beta | 0.2689 | 0.1517 | 0.0282 | 0.2854 | 0.5396 | 1.02 | 41 |
| sigma.y | 0.7504 | 0.0190 | 0.7147 | 0.7500 | 0.7888 | 1.00 | 1891 |
| rho | -0.1768 | 0.4116 | -0.7828 | -0.2527 | 0.9074 | 1.06 | 140 |
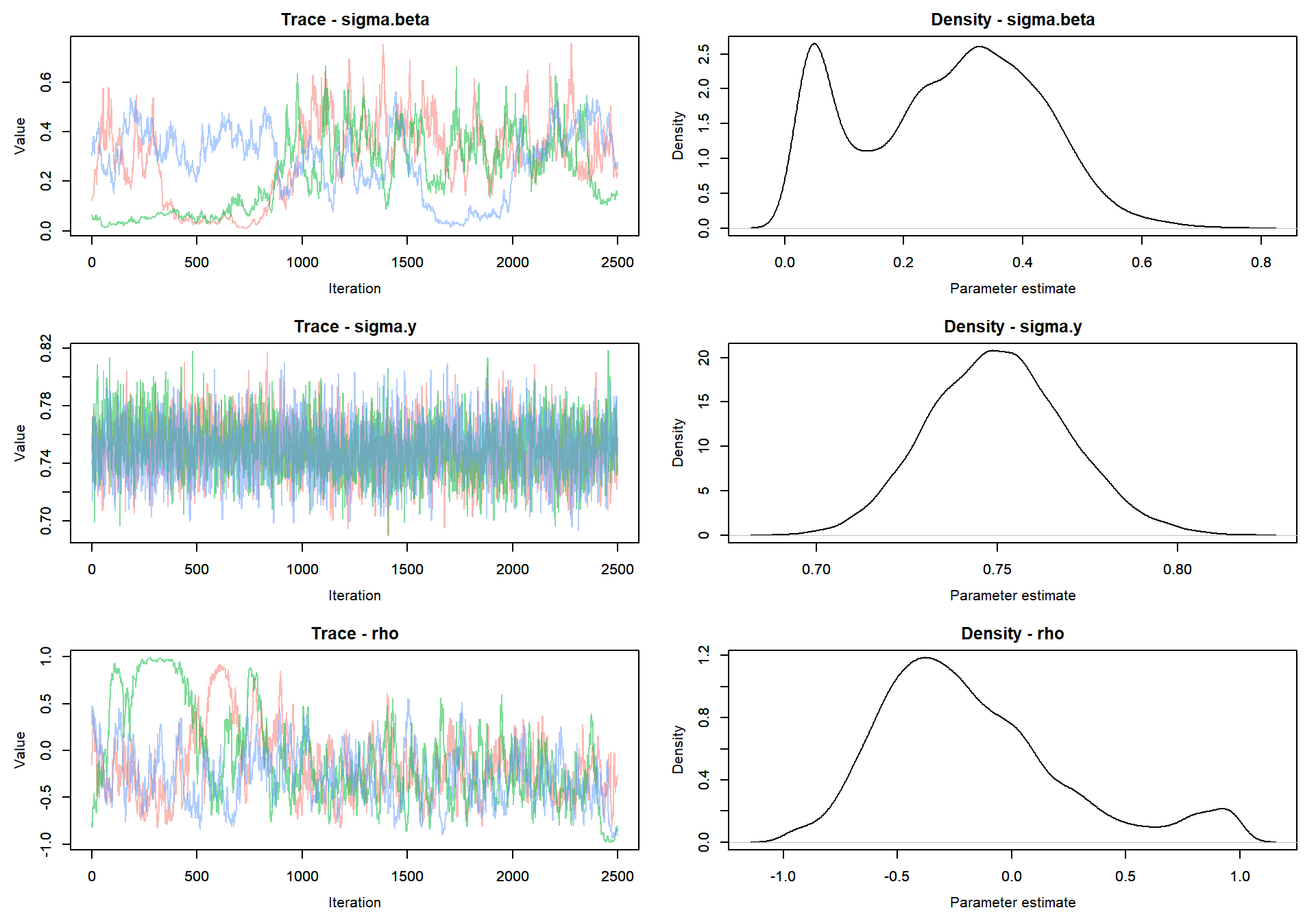
Trace plot of parameters
MCMCvis::MCMCtrace(mlm_intslp_pred, params = params[!params %in% c("alpha", "beta")], pdf = FALSE )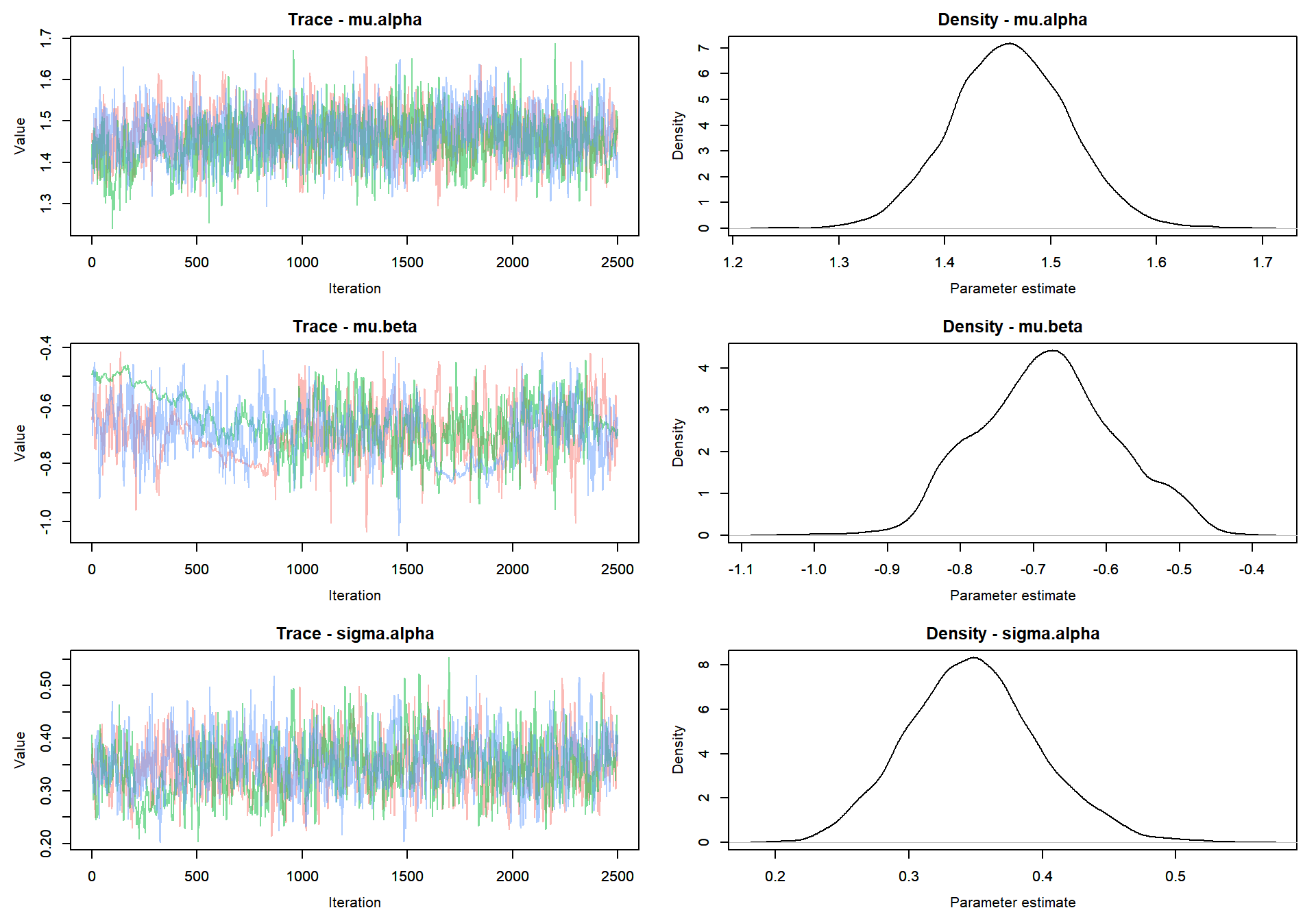
Compare fully Bayesian approach to linear mixed-effects model (via Restricted Maximum Likelihood [REML])
# compare
MCMCvis::MCMCsummary(mlm_intslp_pred, params = "alpha") %>%
data.frame() %>%
dplyr::select("mean") %>%
dplyr::slice_head(n = 8) %>%
dplyr::bind_cols(coef(M3)$county[1:8,"(Intercept)"]) %>%
dplyr::rename(Bayesian=1, LMM=2) %>%
kableExtra::kable(
caption = "alpha predictions Bayesian vs. LMM (lme4::lmer)"
, digits = 4
) %>%
kableExtra::kable_styling(font_size = 12)| Bayesian | LMM | |
|---|---|---|
| alpha[1] | 1.1526 | 1.1445 |
| alpha[2] | 0.9326 | 0.9334 |
| alpha[3] | 1.4738 | 1.4717 |
| alpha[4] | 1.5203 | 1.5354 |
| alpha[5] | 1.4267 | 1.4270 |
| alpha[6] | 1.4805 | 1.4827 |
| alpha[7] | 1.8297 | 1.8194 |
| alpha[8] | 1.6816 | 1.6908 |
# compare
MCMCvis::MCMCsummary(mlm_intslp_pred, params = "beta") %>%
data.frame() %>%
dplyr::select("mean") %>%
dplyr::slice_head(n = 8) %>%
dplyr::bind_cols(coef(M3)$county[1:8,"x"]) %>%
dplyr::rename(Bayesian=1, LMM=2) %>%
kableExtra::kable(
caption = "beta predictions Bayesian vs. LMM (lme4::lmer)"
, digits = 4
) %>%
kableExtra::kable_styling(font_size = 12)| Bayesian | LMM | |
|---|---|---|
| beta[1] | -0.5844 | -0.5406 |
| beta[2] | -0.7544 | -0.7708 |
| beta[3] | -0.6696 | -0.6689 |
| beta[4] | -0.7284 | -0.7526 |
| beta[5] | -0.6362 | -0.6207 |
| beta[6] | -0.6848 | -0.6877 |
| beta[7] | -0.5250 | -0.4748 |
| beta[8] | -0.6849 | -0.6975 |
4.4 Including group-level predictors (p. 280)
We can expand the varying-intercept, varying-slope model including \(x\) of (\(\alpha\) and \(\beta\)) by including a group-level predictor (in this case, soil uranium):
\[\begin{align*} y_i &\sim \textrm{N}(\alpha_{j[i]} + \beta_{j[i]} \cdot x_i, \sigma^2_y) \\ &\textrm{for} \; i = 1, \cdots, n \\ \left(\begin{array}{c} \alpha_{j}\\ \beta_{j} \end{array}\right) &\sim \textrm{N}\left(\left(\begin{array}{c} \mu_\alpha\\ \mu_\beta \end{array}\right) , \left(\begin{array}{cc} \sigma^{2}_{\alpha} & \rho\sigma_{\alpha}\sigma_{\beta}\\ \rho\sigma_{\alpha}\sigma_{\beta} & \sigma^{2}_{\beta} \end{array}\right) \right) \\ &\textrm{where:}\\ &\;\mu_\alpha = \gamma^{\alpha}_{0} + \gamma^{\alpha}_{1} \cdot u_j\\ &\;\mu_\beta = \gamma^{\beta}_{0} + \gamma^{\beta}_{1} \cdot u_j\\ &\textrm{for} \; j = 1, \cdots, J \end{align*}\]
4.4.1 Using lme4::lmer (p. 281)
## Including group level predictors
M4 <- lme4::lmer(data = radon_mn, formula = y ~ x + u + x:u + (1 + x | county))
summary(M4)Linear mixed model fit by REML [‘lmerMod’] Formula: y ~ x + u + x:u + (1 + x | county) Data: radon_mn
REML criterion at convergence: 2132.2
Scaled residuals: Min 1Q Median 3Q Max -5.2770 -0.6057 0.0507 0.6288 3.4495
Random effects:
Groups Name Variance Std.Dev. Corr
county (Intercept) 0.0000 0.0000
x 0.1392 0.3731 NaN
Residual 0.5735 0.7573
Number of obs: 919, groups: county, 85
Fixed effects: Estimate Std. Error t value (Intercept) 1.44407 0.02926 49.353 x -0.64131 0.08915 -7.194 u 0.84628 0.07476 11.320 x:u -0.41582 0.23894 -1.740
Correlation of Fixed Effects:
(Intr) x u
x -0.328
u 0.354 -0.116
x:u -0.111 0.155 -0.313
optimizer (nloptwrap) convergence code: 0 (OK)
boundary (singular) fit: see help(‘isSingular’)
#estimated regression coefficicents
coef(M4)$county[1:3,] (Intercept) x u x:uAITKIN 1.444075 -0.5861689 0.8462757 -0.4158211 ANOKA 1.444075 -0.9559927 0.8462757 -0.4158211 BECKER 1.444075 -0.6121327 0.8462757 -0.4158211
# fixed and random effects
fixef(M4)(Intercept) x u x:u 1.4440747 -0.6413080 0.8462757 -0.4158211
ranef(M4)$county[1:3,] (Intercept) xAITKIN 0 0.05513916 ANOKA 0 -0.31468472 BECKER 0 0.02917531
# check intercept match for county 1
fixef(M4)["(Intercept)"] + fixef(M4)["u"] * u$u[1] + ranef(M4)$county[1,"(Intercept)"](Intercept) 0.8609504
coef(M4)$county[1,"(Intercept)"][1] 1.444075
# check slope match for county 1
fixef(M4)["x"] + fixef(M4)["x:u"] * u$u[1] + ranef(M4)$county[1,"x"] x -0.2996483
coef(M4)$county[1,"x"][1] -0.5861689
print("!!! yoinks! this doesn't align with book but the data and code are the same as the book...maybe bayes can help")[1] “!!! yoinks! this doesn’t align with book but the data and code are the same as the book…maybe bayes can help”
yoinks! this doesn’t align with book but the data and code are the same as the book…maybe bayes can help
4.4.2 Bayesian
4.4.2.1 Data and Initial conditions
# varying-intercept model, no predictors
## data for jags
data <- list(
n = nrow(radon_mn)
, J = radon_mn$county %>% unique() %>% length()
, y = radon_mn$log.radon %>% as.double()
, x = radon_mn$x %>% as.double()
, county = radon_mn$county_index %>% as.double()
, u = u$u %>% as.double()
)
## inits for jags
inits <- function(){
list(
B.matrix = array(data = rnorm(n=2*data$J, mean = 0, sd = 1), dim = c(data$J,2)) # 2 = num. model params
# mu.alpha
, gamma.alpha0 = rnorm(1, mean = 0, sd = 1)
, gamma.alpha1 = rnorm(1, mean = 0, sd = 1)
# mu.beta
, gamma.beta0 = rnorm(1, mean = 0, sd = 1)
, gamma.beta1 = rnorm(1, mean = 0, sd = 1)
, sigma.alpha = runif(n = 1, min = 0, max = 1)
, sigma.beta = runif(n = 1, min = 0, max = 1)
, sigma.y = runif(n = 1, min = 0, max = 1)
, rho = runif(n = 1, min = -1, max = 1)
)
}
# define parameters to return from MCMC
params <- c (
"alpha"
, "beta"
# mu.alpha
, "gamma.alpha0"
, "gamma.alpha1"
# mu.beta
, "gamma.beta0"
, "gamma.beta1"
, "sigma.alpha"
, "sigma.beta"
, "sigma.y"
, "rho"
)4.4.2.2 JAGS Model
Recall model defined above
Write out the JAGS code for the model.
## JAGS Model
model {
###########################
# priors
###########################
# coefficient of correlation for covariance matrix
rho ~ dunif(-1,1)
# alpha priors
sigma.alpha ~ dunif(0, 100)
tau.alpha <- 1/sigma.alpha^2
# mu.alpha
gamma.alpha0 ~ dnorm(0, .0001)
gamma.alpha1 ~ dnorm(0, .0001)
# beta priors
sigma.beta ~ dunif(0, 100)
tau.beta <- 1/sigma.beta^2
# mu.beta
gamma.beta0 ~ dnorm(0, .0001)
gamma.beta1 ~ dnorm(0, .0001)
# y priors
sigma.y ~ dunif(0, 100)
tau.y <- 1/sigma.y^2
###########################
# variance-covariance matrix
###########################
# variance
Sigma.matrix[1, 1] <- sigma.alpha^2 # row 1, column 1
Sigma.matrix[2, 2] <- sigma.beta^2 # row 2, column 2
# Cov(alpha,beta) = rho * sigma.alpha * sigma.beta
Sigma.matrix[2, 1] <- rho * sigma.alpha * sigma.beta # row 2, column 1
Sigma.matrix[1, 2] <- Sigma.matrix[2, 1] # row 1, column 2
# invert Sigma matrix
Omega.matrix <- inverse(Sigma.matrix)
###########################
# likelihood
###########################
# varying slopes and intercepts at the county level
for(j in 1:J){
mu.alpha[j] <- gamma.alpha0 + gamma.alpha1 * u[j]
mu.beta[j] <- gamma.beta0 + gamma.beta1 * u[j]
# put mu.alpha and mu.beta in matrix
B.hat[j,1] <- mu.alpha[j] # column 1
B.hat[j,2] <- mu.beta[j] # column 2
# note multivariate normal
B.matrix[j,1:2] ~ dmnorm(B.hat[j,], Omega.matrix)
# extract alpha and beta from matrix
alpha[j] <- B.matrix[j, 1] # column 1
beta[j] <- B.matrix[j, 2] # column 2
}
# y
for (i in 1:n){
mu.y[i] <- alpha[county[i]] + beta[county[i]] * x[i]
y[i] ~ dnorm(mu.y[i], tau.y)
}
}4.4.2.3 Implement JAGS Model
##################################################################
# insert JAGS model code into an R script
##################################################################
{ # Extra bracket needed only for R markdown files - see answers
sink("intslp_grp_pred.R") # This is the file name for the jags code
cat("
## JAGS Model
model {
###########################
# priors
###########################
# coefficient of correlation for covariance matrix
rho ~ dunif(-1,1)
# alpha priors
sigma.alpha ~ dunif(0, 100)
tau.alpha <- 1/sigma.alpha^2
# mu.alpha
gamma.alpha0 ~ dnorm(0, .0001)
gamma.alpha1 ~ dnorm(0, .0001)
# beta priors
sigma.beta ~ dunif(0, 100)
tau.beta <- 1/sigma.beta^2
# mu.beta
gamma.beta0 ~ dnorm(0, .0001)
gamma.beta1 ~ dnorm(0, .0001)
# y priors
sigma.y ~ dunif(0, 100)
tau.y <- 1/sigma.y^2
###########################
# variance-covariance matrix
###########################
# variance
Sigma.matrix[1, 1] <- sigma.alpha^2 # row 1, column 1
Sigma.matrix[2, 2] <- sigma.beta^2 # row 2, column 2
# Cov(alpha,beta) = rho * sigma.alpha * sigma.beta
Sigma.matrix[2, 1] <- rho * sigma.alpha * sigma.beta # row 2, column 1
Sigma.matrix[1, 2] <- Sigma.matrix[2, 1] # row 1, column 2
# invert Sigma matrix
Omega.matrix <- inverse(Sigma.matrix)
###########################
# likelihood
###########################
# varying slopes and intercepts at the county level
for(j in 1:J){
mu.alpha[j] <- gamma.alpha0 + gamma.alpha1 * u[j]
mu.beta[j] <- gamma.beta0 + gamma.beta1 * u[j]
# put mu.alpha and mu.beta in matrix
B.hat[j,1] <- mu.alpha[j] # column 1
B.hat[j,2] <- mu.beta[j] # column 2
# note multivariate normal
B.matrix[j,1:2] ~ dmnorm(B.hat[j,], Omega.matrix)
# extract alpha and beta from matrix
alpha[j] <- B.matrix[j, 1] # column 1
beta[j] <- B.matrix[j, 2] # column 2
}
# y
for (i in 1:n){
mu.y[i] <- alpha[county[i]] + beta[county[i]] * x[i]
y[i] ~ dnorm(mu.y[i], tau.y)
}
}
", fill = TRUE)
sink()
}
##################################################################
# implement model
##################################################################
######################
# Call to JAGS
######################
intslp_grp_pred = rjags::jags.model(
file = "intslp_grp_pred.R"
, data = data
, inits = inits
, n.chains = 3
, n.adapt = 1000
)## Compiling model graph
## Resolving undeclared variables
## Allocating nodes
## Graph information:
## Observed stochastic nodes: 919
## Unobserved stochastic nodes: 93
## Total graph size: 3837
##
## Initializing modelstats::update(intslp_grp_pred, n.iter = 1000, progress.bar = "none")
# save the coda object (more precisely, an mcmc.list object) to R
mlm_intslp_grp_pred = rjags::coda.samples(
model = intslp_grp_pred
, variable.names = params
, n.iter = 5000
, n.thin = 1
, progress.bar = "none"
)4.4.2.4 Summary of the marginal posterior distributions of the parameters
Compare to results from scaled inverse-Wishart JAGS model results
# summary
MCMCvis::MCMCsummary(mlm_intslp_grp_pred, params = params[!params %in% c("alpha", "beta")]) %>%
kableExtra::kable(
caption = "Bayesian: Varying-intercept & slope multilevel model, floor of measurement predictor + county-level (grp) uranium"
, digits = 4
) %>%
kableExtra::kable_styling(font_size = 12)| mean | sd | 2.5% | 50% | 97.5% | Rhat | n.eff | |
|---|---|---|---|---|---|---|---|
| gamma.alpha0 | 1.4701 | 0.0363 | 1.3997 | 1.4702 | 1.5422 | 1.00 | 779 |
| gamma.alpha1 | 0.8075 | 0.0941 | 0.6280 | 0.8071 | 0.9931 | 1.02 | 914 |
| gamma.beta0 | -0.6853 | 0.0888 | -0.8510 | -0.6886 | -0.5007 | 1.00 | 569 |
| gamma.beta1 | -0.4326 | 0.2414 | -0.9258 | -0.4300 | 0.0298 | 1.02 | 497 |
| sigma.alpha | 0.1401 | 0.0476 | 0.0571 | 0.1375 | 0.2444 | 1.01 | 233 |
| sigma.beta | 0.3009 | 0.1170 | 0.0850 | 0.2970 | 0.5389 | 1.01 | 188 |
| sigma.y | 0.7513 | 0.0185 | 0.7151 | 0.7511 | 0.7878 | 1.00 | 4958 |
| rho | 0.2917 | 0.4392 | -0.5959 | 0.3146 | 0.9668 | 1.00 | 183 |
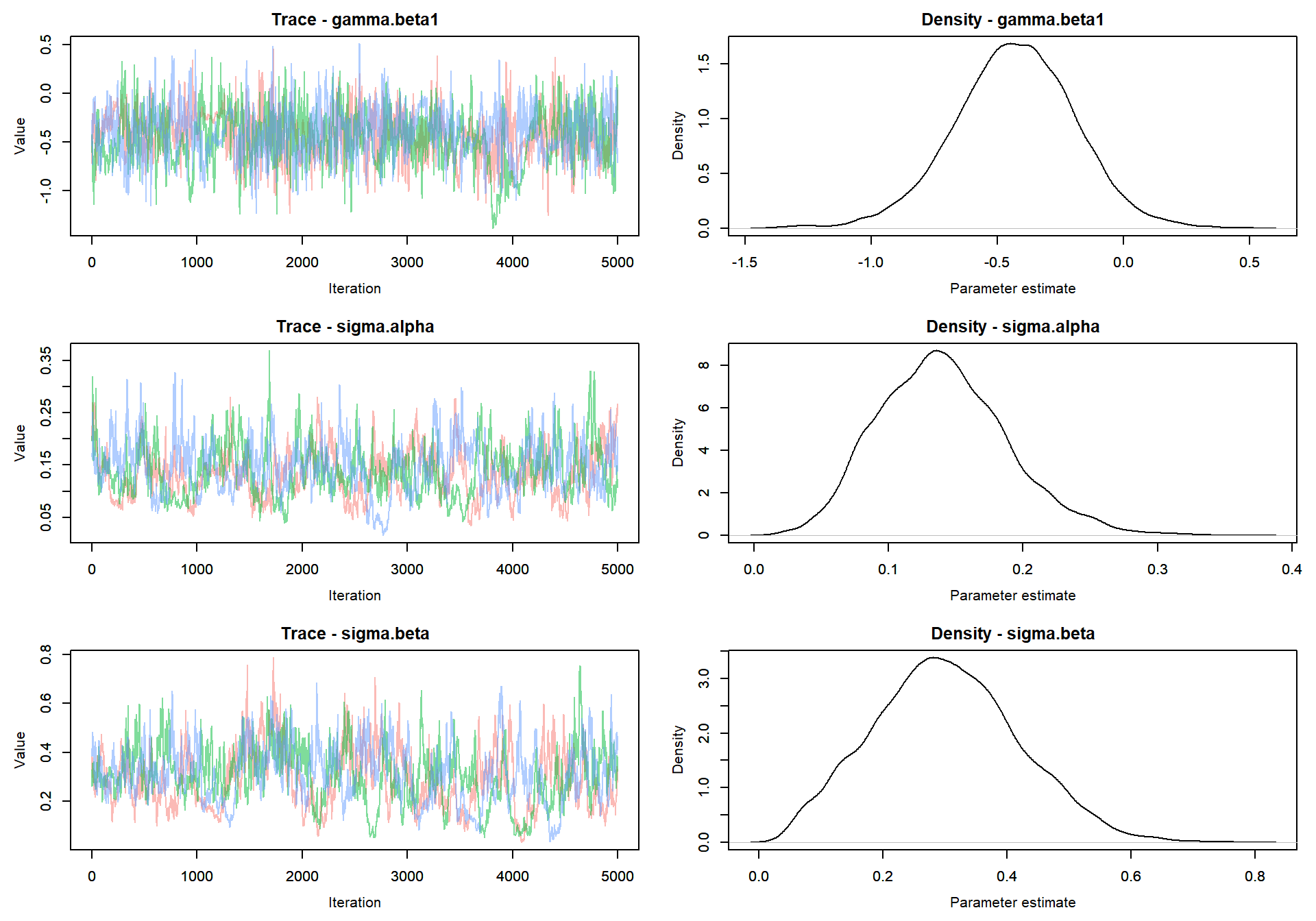
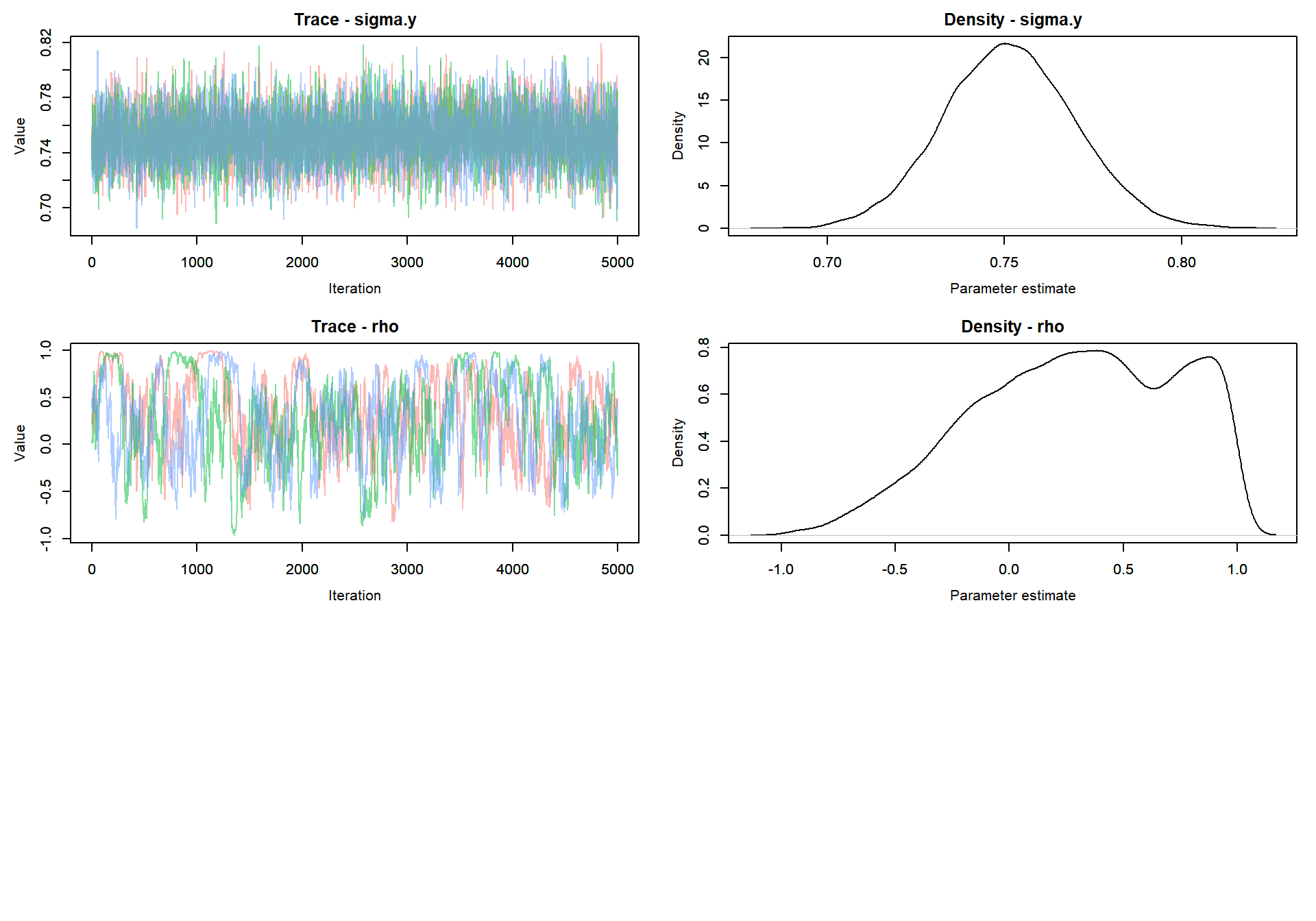
Trace plot of parameters
MCMCvis::MCMCtrace(mlm_intslp_grp_pred, params = params[!params %in% c("alpha", "beta")], pdf = FALSE )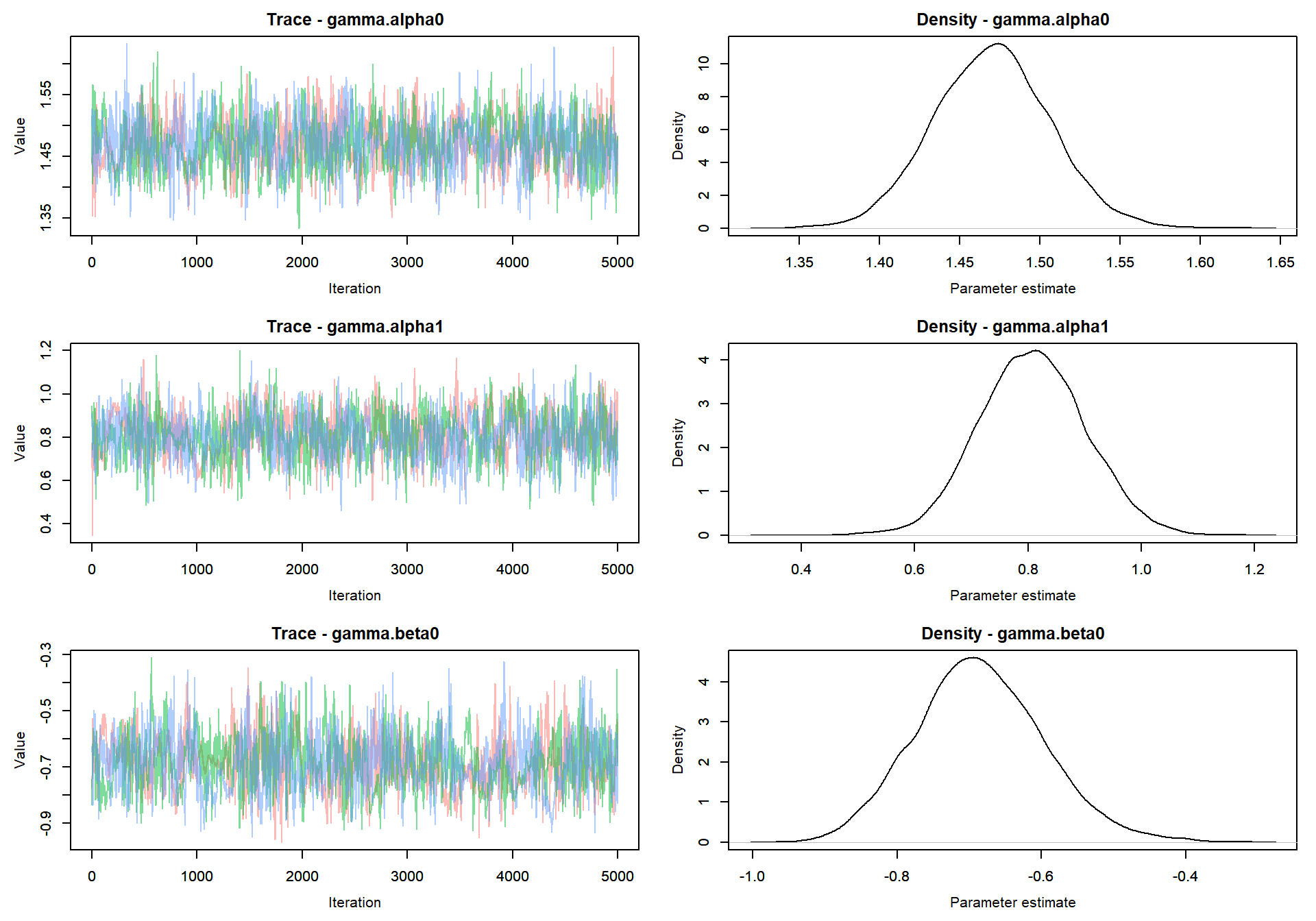
Caterpillar plot of parameters
# Caterpillar plots
MCMCvis::MCMCplot(mlm_intslp_grp_pred, params = params[!params %in% c("alpha", "beta")], xlim = c(-1,2))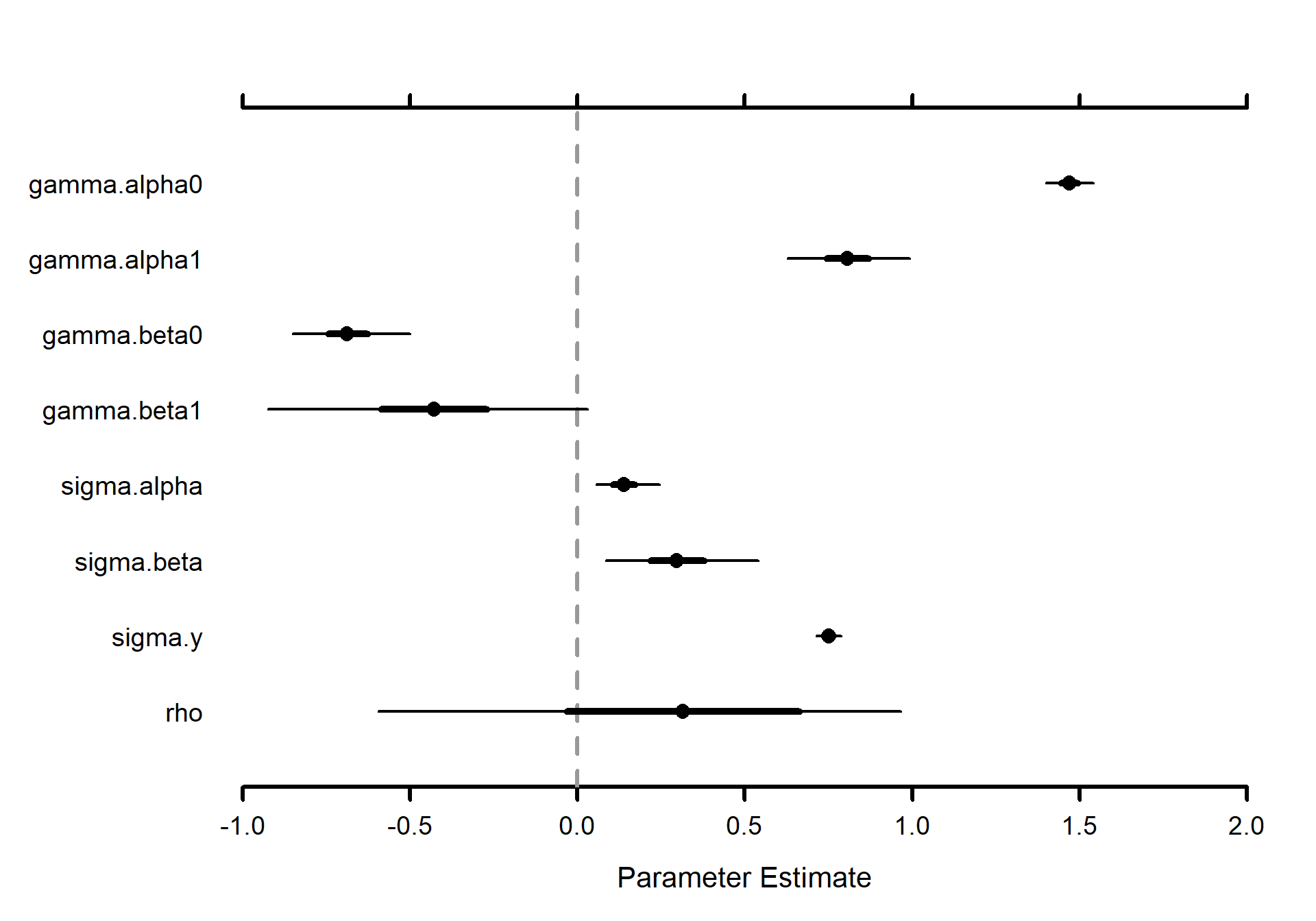
Compare fully Bayesian approach to linear mixed-effects model (via Restricted Maximum Likelihood [REML])
# compare
MCMCvis::MCMCsummary(mlm_intslp_grp_pred, params = "alpha") %>%
data.frame() %>%
dplyr::select("mean") %>%
dplyr::slice_head(n = 8) %>%
dplyr::bind_cols(coef(M4)$county[1:8,"(Intercept)"]) %>%
dplyr::rename(Bayesian=1, LMM=2) %>%
kableExtra::kable(
caption = "alpha predictions Bayesian vs. LMM (lme4::lmer)"
, digits = 4
) %>%
kableExtra::kable_styling(font_size = 12)| Bayesian | LMM | |
|---|---|---|
| alpha[1] | 0.8954 | 1.4441 |
| alpha[2] | 0.8180 | 1.4441 |
| alpha[3] | 1.3888 | 1.4441 |
| alpha[4] | 1.0627 | 1.4441 |
| alpha[5] | 1.3640 | 1.4441 |
| alpha[6] | 1.7555 | 1.4441 |
| alpha[7] | 1.8028 | 1.4441 |
| alpha[8] | 1.7393 | 1.4441 |
# compare
MCMCvis::MCMCsummary(mlm_intslp_grp_pred, params = "beta") %>%
data.frame() %>%
dplyr::select("mean") %>%
dplyr::slice_head(n = 8) %>%
dplyr::bind_cols(coef(M4)$county[1:8,"x"]) %>%
dplyr::rename(Bayesian=1, LMM=2) %>%
kableExtra::kable(
caption = "beta predictions Bayesian vs. LMM (lme4::lmer)"
, digits = 4
) %>%
kableExtra::kable_styling(font_size = 12)| Bayesian | LMM | |
|---|---|---|
| beta[1] | -0.3568 | -0.5862 |
| beta[2] | -0.5320 | -0.9560 |
| beta[3] | -0.6137 | -0.6121 |
| beta[4] | -0.3763 | -0.5563 |
| beta[5] | -0.5779 | -0.5781 |
| beta[6] | -0.8663 | -0.6413 |
| beta[7] | -0.4844 | -0.2199 |
| beta[8] | -0.7457 | -0.5665 |
4.4.2.5 Estimated \(\alpha_{j}\) versus uranium measurment
Replicate Figure 13.2a for the multilevel model where the intercept and the slope vary by county (p.281)
dta_temp <- dplyr::bind_cols(
u
, MCMCvis::MCMCpstr(mlm_intslp_grp_pred, params = "alpha", func = function(x) quantile(x, c(0.25, 0.5, 0.75))) %>%
data.frame() %>%
dplyr::rename_with(
~ tolower(gsub(".", "", .x, fixed = TRUE))
)
) %>%
dplyr::arrange(county_index)
ggplot(dta_temp, mapping = aes(x = u)) +
geom_abline(
intercept = MCMCvis::MCMCpstr(mlm_intslp_grp_pred, params = "gamma.alpha0", func = median) %>% unlist()
, slope = MCMCvis::MCMCpstr(mlm_intslp_grp_pred, params = "gamma.alpha1", func = median) %>% unlist()
, linetype = "dashed"
, lwd = 1
, color = "gray40"
) +
geom_linerange(
mapping = aes(ymin = alpha25, ymax = alpha75)
) +
geom_point(
mapping = aes(y = alpha50)
) +
# scale_y_continuous(limits = c(0,2.5)) +
ylab(latex2exp::TeX("$\\hat{\\alpha}_{j}$")) +
xlab("log(u)") +
labs(
title = latex2exp::TeX("Median model intercept ($\\hat{\\alpha}_{j}$) prediction for each county (j) versus county-level uranium measurement")
) +
theme_bw() +
theme(
axis.text.x = element_blank()
, axis.ticks.x = element_blank()
, panel.grid = element_blank()
)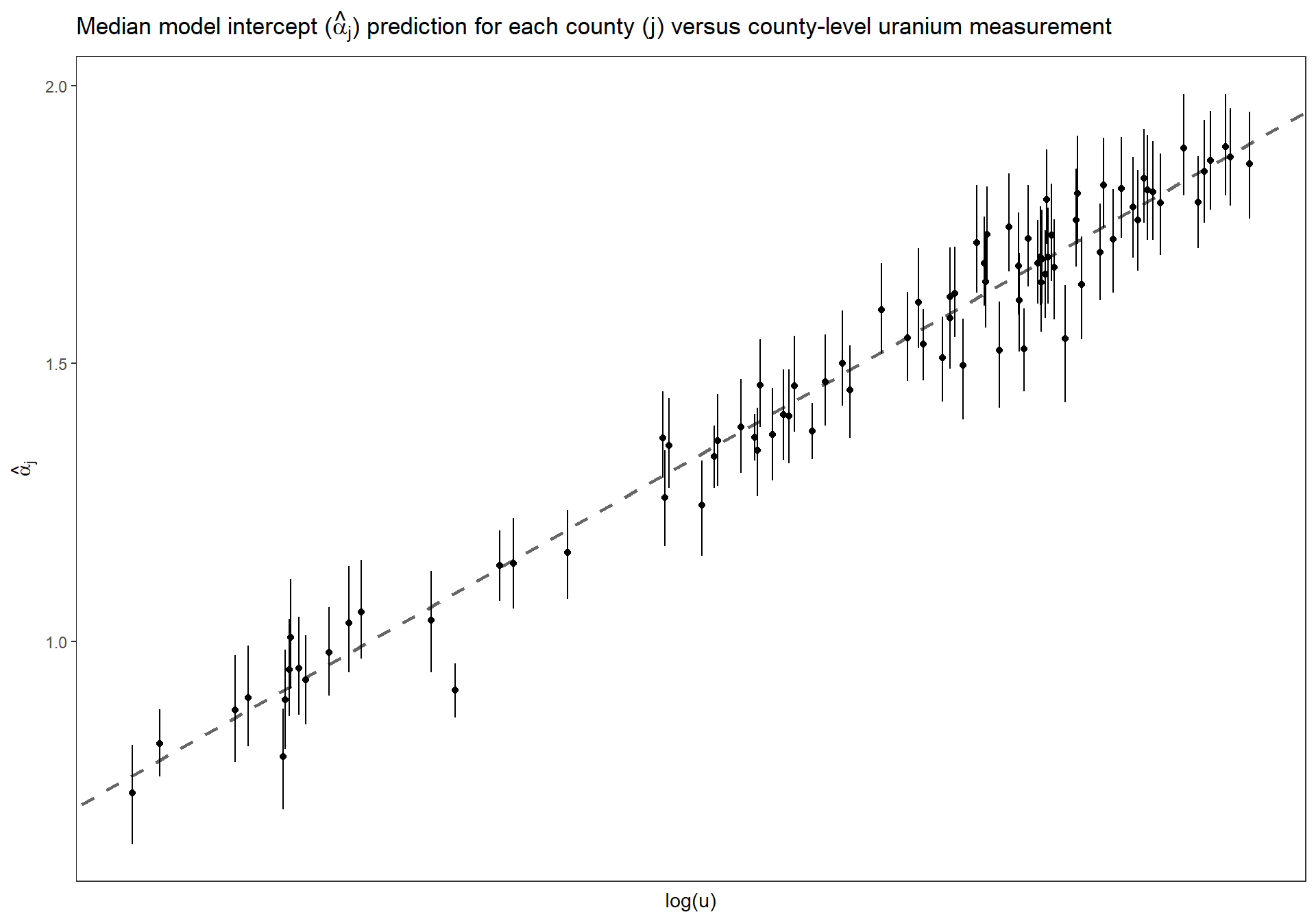
Estimated county coefficients \(\alpha_j\) plotted versus county-level uranium measurement \(u_j\), along with the estimated multilevel regression line \(\alpha_j = \gamma^{\alpha}_{0} + \gamma^{\alpha}_{1} \cdot u_j\). The county coefficients roughly follow the line but not exactly; the deviation of the coefficients from the line is captured in \(\sigma_{\alpha}\), the standard deviation of the errors in the county-level regression.
Replicate Figure 13.2b for the multilevel model where the intercept and the slope vary by county (p.281)
dta_temp <- dplyr::bind_cols(
u
, MCMCvis::MCMCpstr(mlm_intslp_grp_pred, params = "beta", func = function(x) quantile(x, c(0.25, 0.5, 0.75))) %>%
data.frame() %>%
dplyr::rename_with(
~ tolower(gsub(".", "", .x, fixed = TRUE))
)
) %>%
dplyr::arrange(county_index)
ggplot(dta_temp, mapping = aes(x = u)) +
geom_abline(
intercept = MCMCvis::MCMCpstr(mlm_intslp_grp_pred, params = "gamma.beta0", func = median) %>% unlist()
, slope = MCMCvis::MCMCpstr(mlm_intslp_grp_pred, params = "gamma.beta1", func = median) %>% unlist()
, linetype = "dashed"
, lwd = 1
, color = "gray40"
) +
geom_linerange(
mapping = aes(ymin = beta25, ymax = beta75)
) +
geom_point(
mapping = aes(y = beta50)
) +
# scale_y_continuous(limits = c(0,2.5)) +
ylab(latex2exp::TeX("$\\hat{\\beta}_{j}$")) +
xlab("log(u)") +
labs(
title = latex2exp::TeX("Median model slope ($\\hat{\\beta}_{j}$) prediction for each county (j) versus county-level uranium measurement")
) +
theme_bw() +
theme(
axis.text.x = element_blank()
, axis.ticks.x = element_blank()
, panel.grid = element_blank()
)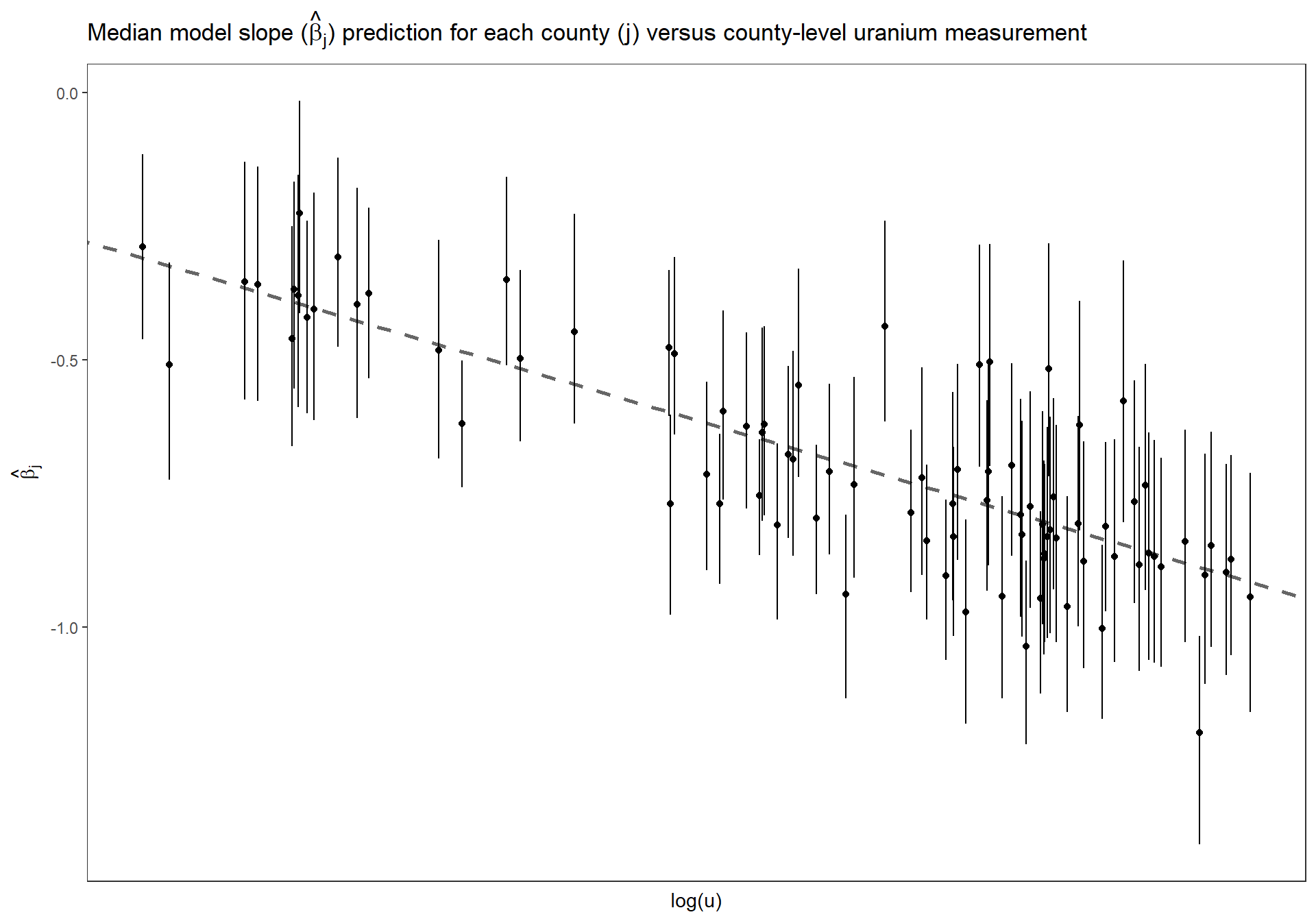
Estimated county coefficients \(\beta_j\) plotted versus county-level uranium measurement \(u_j\), along with the estimated multilevel regression line \(\beta_j = \gamma^{\beta}_{0} + \gamma^{\beta}_{1} \cdot u_j\). The county coefficients roughly follow the line but not exactly; the deviation of the coefficients from the line is captured in \(\sigma_{\beta}\), the standard deviation of the errors in the county-level regression.
4.5 Modeling multiple varying coefficients
When more than two coefficients vary (for example, \(y_i \sim \textrm{N}(\beta_{0} + \beta_{1} \cdot x_{i1} + \beta_{2} \cdot x_{i2}, \sigma^2_y)\) ), with \(\beta_0\), \(\beta_1\), and \(\beta_2\) varying by group), it is helpful to move to matrix notation (upper-case notation) in modeling the coefficients and their group-level regression model and covariance matrix (p.284)
More generally, we can have \(J\) groups, \(K\) individual-level predictors, and \(L\) predictors in the group-level regression (including the constant term as a predictor in both cases). For example, \(K = L = 2\) in the radon model that has floor as an individual predictor and uranium as a county-level predictor. To include group-level predictors (p.285):
\[\begin{align*} y_i &\sim \textrm{N}(X_{i} B_{j[i]}, \sigma^2_y) \\ &\textrm{for} \; i = 1, \cdots, n \\ B_j &\sim \textrm{N}(U_{j} G, \Sigma_B) \\ &\textrm{for} \; j = 1, \cdots, J \end{align*}\]
where
- \(B\) is the \(J \times K\) matrix of individual-level coefficients,
- \(U\) is the \(J \times L\) matrix of group-level predictors (including the constant term), and
- \(G\) is the \(L \times K\) matrix of coefficients for the group-level regression.
- \(U_j\) is the \(j^{th}\) row of \(U\), the vector of predictors for group \(j\), and so \(U_jG\) is a vector of length \(K\)
- \(X\) is the \(n \times K\) matrix of predictors (e.g. \(x_{i1},x_{i2}, \cdots, x_{iK}\)), where \(K\) is the number of individual-level predictors (including the intercept) that vary by group. \(X_i\) is then the vector of length \(K\) representing the \(i^{th}\) row of \(X\)
When more than two coefficients are varying (for example, a varying intercept and two or more varying slopes), it is difficult to model the group-level correlations directly because of constraints involved in the requirement that the covariance matrix be positive definite. With more than two varying coefficients, it is simplest to just use the scaled inverse-Wishart model described above (p.286), using a full matrix notation to allow for an arbitrary number \(K\) of coefficients that vary by group (p.377)
4.5.1 JAGS Model (no group-level predictors; p.378)
Write out the JAGS code for the model.
A useful trick with the scaling is to define the coefficients \(\alpha_j\) , \(\beta_j\) in terms of multiplicative factors, \(\xi_\alpha\), \(\xi_\beta\), with unscaled parameters \(\alpha^{raw}_j\) , \(\beta^{raw}_j\) having the inverse-Wishart distribution.
model {
###########################
# In R
###########################
# J <- data$group_index %>% unique() %>% length()
# X <- cbind(1,x)
# K <- ncol(X)
# W <- diag(K) # (KxK matrix with 1's on diagonal)
# inits <- function(){
# list(
# B.raw = array(data = rnorm(n = J*K), dim = c(J,K))
# , mu.raw = rnorm(n = K)
# , sigma.y = runif(n = 1)
# , Tau.B.raw = rWishart(n = 1, df = K+1, Sigma = diag(K))
# , xi = runif(K)
# )
# }
# # parameters of interest
# parameters <- c ("B", "mu", "sigma.y", "sigma.B", "rho.B")
###########################
# priors
###########################
########
# y priors
########
sigma.y ~ dunif(0, 100)
tau.y <- 1/sigma.y^2
########
# define the coefficients for the intercept (e.g. alpha) and slope (e.g. beta)
# in terms of multiplicative factors $\xi_\alpha$, $\xi_\beta$
# with unscaled parameters $\alpha^{raw}_j$ , $\beta^{raw}_j$
########
for(k in 1:K){
xi[k] ~ dunif(0, 100)
mu.raw[k] ~ dnorm(0, .0001)
# parameter means (e.g. mu_alpha and mu_beta)
mu[k] <- xi[k]*mu.raw[k]
}
########
# Specification of the Wishart distribution
########
# for the inverse-variance Tau.B.raw
# , the matrix W is the prior scale
# , which we set to the identity matrix (in R, we write W <- diag(K))
# , and the degrees of freedom set to 1 more than the dimension of the matrix
# (to induce a uniform prior distribution on rho)
df <- K+1
Tau.B.raw[1:K,1:K] ~ dwish(W[,], df)
Sigma.B.raw[1:K,1:K] <- inverse(Tau.B.raw[,])
# loop over K number of individual-level predictors
# (including the intercept) that vary by group
for(k in 1:K){
for(k.prime in 1:K){
# coefficient of correlation (rho) for parameters
rho.B[k,k.prime] <- Sigma.B.raw[k,k.prime] /
sqrt(Sigma.B.raw[k,k]*Sigma.B.raw[k.prime,k.prime])
}
# variance for the intercept and slope parameters
sigma.B[k] <- abs(xi[k])*sqrt(Sigma.B.raw[k,k])
}
###########################
# likelihood
###########################
# varying slopes and intercepts
for(j in 1:J){
for(k in 1:K){
# estimated slope and intercept coefficients
B[j,k] <- xi[k]*B.raw[j,k]
}
B.raw[j,1:K] ~ dmnorm(mu.raw[], Tau.B.raw[,])
}
# y
for(i in 1:n){
# mu.y[i] <- alpha[county[i]] + beta[county[i]] * x[i]
mu.y[i] <- inprod(B[county[i],],X[i,])
y[i] ~ dnorm(mu.y[i], tau.y)
}
}4.5.2 JAGS Model (yes group-level predictors; p.379)
Multiple varying coefficients with multiple group-level predictors
Write out the JAGS code for the model.
This model is analogous to this model above but extends to models with multiple varying coefficients
model {
###########################
# In R
###########################
# J <- data$group_index %>% unique() %>% length()
# X <- cbind(1,x) # x is of length i,...,n
# K <- ncol(X)
# W <- diag(K) # (KxK matrix with 1's on diagonal)
# U <- cbind(1,u) # u is of length j,...,J
# L <- ncol(U)
# inits <- function(){
# list(
# B.raw = array(data = rnorm(n = J*K), dim = c(J,K))
# , G.raw = array(data = rnorm(n = L*K), dim = c(L,K))
# , sigma.y = runif(n = 1)
# , Tau.B.raw = rWishart(n = 1, df = K+1, Sigma = diag(K))
# , xi = runif(K)
# )
# }
# # parameters of interest
# parameters <- c ("B", "G", "sigma.y", "sigma.B", "rho.B")
###########################
# priors
###########################
########
# y priors
########
sigma.y ~ dunif(0, 100)
tau.y <- 1/sigma.y^2
########
# Modeling multiple group-level predictors
########
# If we have several group-level predictors (expressed as a matrix U with J rows, one
# for each group), we use notation such as
for (k in 1:K){
for (l in 1:L){
# parameter means (e.g. gamma.alpha0, gamma.alpha1, gamma.beta0, gamma.beta1)
G[k,l] <- xi[k]*G.raw[k,l]
G.raw[k,l] ~ dnorm(0, .0001)
}
xi[k] ~ dunif(0, 100)
}
########
# Specification of the Wishart distribution
########
# for the inverse-variance Tau.B.raw
# , the matrix W is the prior scale
# , which we set to the identity matrix (in R, we write W <- diag(K))
# , and the degrees of freedom set to 1 more than the dimension of the matrix
# (to induce a uniform prior distribution on rho)
df <- K+1
Tau.B.raw[1:K,1:K] ~ dwish(W[,], df)
Sigma.B.raw[1:K,1:K] <- inverse(Tau.B.raw[,])
# loop over K number of individual-level predictors
# (including the intercept) that vary by group
for(k in 1:K){
for(k.prime in 1:K){
# coefficient of correlation (rho) for parameters
rho.B[k,k.prime] <- Sigma.B.raw[k,k.prime] /
sqrt(Sigma.B.raw[k,k]*Sigma.B.raw[k.prime,k.prime])
}
# variance for the intercept and slope parameters
sigma.B[k] <- abs(xi[k])*sqrt(Sigma.B.raw[k,k])
}
########
# Modeling multiple varying coefficients
########
# Modeling multiple varying coefficients
for (j in 1:J){
B.raw[j,1:K] ~ dmnorm(B.raw.hat[j,], Tau.B.raw[,])
for (k in 1:K){
# parameter means (e.g. mu.alpha[j] <- gamma.alpha0 + gamma.alpha1 * u[j])
B.raw.hat[j,k] <- inprod(G.raw[k,],U[j,])
}
}
###########################
# likelihood
###########################
# varying slopes and intercepts
for (k in 1:K){
for (j in 1:J){
# estimated slope and intercept coefficients
B[j,k] <- xi[k]*B.raw[j,k]
}
}
# y
for(i in 1:n){
# mu.y[i] <- alpha[county[i]] + beta[county[i]] * x[i]
mu.y[i] <- inprod(B[county[i],],X[i,])
y[i] ~ dnorm(mu.y[i], tau.y)
}
}4.5.2.1 Data and Initial conditions
# select predictor variables for y model
x_vars_nms <- c("intercept", "x")
x_vars <- radon_mn %>%
dplyr::mutate(intercept = 1) %>%
dplyr::select(dplyr::any_of(x_vars_nms)) %>%
dplyr::mutate_if(is.numeric, as.double)
# select predictor variables for group-level model
grp_vars_nms <- c("intercept", "u")
grp_vars <- radon_mn %>%
dplyr::mutate(intercept = 1) %>%
dplyr::group_by(county_index) %>%
dplyr::summarise_at(grp_vars_nms, dplyr::first) %>%
dplyr::ungroup() %>%
dplyr::arrange(county_index) %>%
dplyr::select(dplyr::any_of(grp_vars_nms)) %>%
dplyr::mutate_if(is.numeric, as.double)
# set up matrix form
J_temp <- radon_mn$county %>% unique() %>% length()
# X_temp <- cbind(1, x_vars) # x_vars is of length i,...,n
X_temp <- x_vars # added intercept above in data definition
K_temp <- ncol(X_temp)
W_temp <- diag(K_temp) # (KxK matrix with 1's on diagonal)
# U_temp <- cbind(1, grp_vars) # grp_vars is of length j,...,J
U_temp <- grp_vars # added intercept above in data definition
L_temp <- ncol(U_temp)
## data for jags
data <- list(
n = nrow(radon_mn)
, y = radon_mn$log.radon %>% as.double()
, county = radon_mn$county_index %>% as.double()
# matrix form
, J = J_temp
, X = X_temp
, K = K_temp
, W = W_temp
, U = U_temp
, L = L_temp
)
## inits for jags
inits <- function(){
list(
B.raw = array(data = rnorm(n = data$J*data$K, mean = 0, sd = 1), dim = c(data$J, data$K))
, G.raw = array(data = rnorm(n = data$L*data$K), dim = c(data$L, data$K))
, sigma.y = runif(n = 1, min = 0, max = 1)
# , Tau.B.raw = rWishart(n = 1, df = data$K+1, Sigma = diag(data$K))
, Tau.B.raw = monomvn::rwish(v = data$K+1, S = diag(data$K))
, xi = runif(n = data$K, min = 0, max = 1)
)
}
# define parameters to return from MCMC
params <- c (
"B"
, "G"
, "sigma.y"
, "sigma.B"
, "rho.B"
)4.5.2.2 Implement JAGS Model
##################################################################
# insert JAGS model code into an R script
##################################################################
{ # Extra bracket needed only for R markdown files - see answers
sink("intslp_grp_pred_mtrx.R") # This is the file name for the jags code
cat("
model {
###########################
# priors
###########################
########
# y priors
########
sigma.y ~ dunif(0, 100)
tau.y <- 1/sigma.y^2
########
# Modeling multiple group-level predictors
########
# If we have several group-level predictors (expressed as a matrix U with J rows, one
# for each group), we use notation such as
for (k in 1:K){
for (l in 1:L){
# parameter means (e.g. gamma.alpha0, gamma.alpha1, gamma.beta0, gamma.beta1)
G[k,l] <- xi[k]*G.raw[k,l]
G.raw[k,l] ~ dnorm(0, .0001)
}
xi[k] ~ dunif(0, 100)
}
########
# Specification of the Wishart distribution
########
# for the inverse-variance Tau.B.raw
# , the matrix W is the prior scale
# , which we set to the identity matrix (in R, we write W <- diag(K))
# , and the degrees of freedom set to 1 more than the dimension of the matrix
# (to induce a uniform prior distribution on rho)
df <- K+1
Tau.B.raw[1:K,1:K] ~ dwish(W[,], df)
Sigma.B.raw[1:K,1:K] <- inverse(Tau.B.raw[,])
# loop over K number of individual-level predictors
# (including the intercept) that vary by group
for(k in 1:K){
for(k.prime in 1:K){
# coefficient of correlation (rho) for parameters
rho.B[k,k.prime] <- Sigma.B.raw[k,k.prime] /
sqrt(Sigma.B.raw[k,k]*Sigma.B.raw[k.prime,k.prime])
}
# variance for the intercept and slope parameters
sigma.B[k] <- abs(xi[k])*sqrt(Sigma.B.raw[k,k])
}
########
# Modeling multiple varying coefficients
########
# Modeling multiple varying coefficients
for (j in 1:J){
B.raw[j,1:K] ~ dmnorm(B.raw.hat[j,], Tau.B.raw[,])
for (k in 1:K){
# parameter means (e.g. mu.alpha[j] <- gamma.alpha0 + gamma.alpha1 * u[j])
B.raw.hat[j,k] <- inprod(G.raw[k,],U[j,])
}
}
###########################
# likelihood
###########################
# varying slopes and intercepts
for (k in 1:K){
for (j in 1:J){
# estimated slope and intercept coefficients
B[j,k] <- xi[k]*B.raw[j,k]
}
}
# y
for(i in 1:n){
# mu.y[i] <- alpha[county[i]] + beta[county[i]] * x[i]
mu.y[i] <- inprod(B[county[i],],X[i,])
y[i] ~ dnorm(mu.y[i], tau.y)
}
}
", fill = TRUE)
sink()
}
##################################################################
# implement model
##################################################################
######################
# Call to JAGS
######################
intslp_grp_pred_mtrx = rjags::jags.model(
file = "intslp_grp_pred_mtrx.R"
, data = data
, inits = inits
, n.chains = 3
, n.adapt = 1000
)## Compiling model graph
## Resolving undeclared variables
## Allocating nodes
## Graph information:
## Observed stochastic nodes: 919
## Unobserved stochastic nodes: 93
## Total graph size: 5814
##
## Initializing modelstats::update(intslp_grp_pred_mtrx, n.iter = 1000, progress.bar = "none")
# save the coda object (more precisely, an mcmc.list object) to R
mlm_intslp_grp_pred_mtrx = rjags::coda.samples(
model = intslp_grp_pred_mtrx
, variable.names = params
, n.iter = 5000
, n.thin = 1
, progress.bar = "none"
)4.5.2.3 Summary of the marginal posterior distributions of the parameters
Compare to results from multivariate normal JAGS model results where \(K = L = 2\)
# summary
MCMCvis::MCMCsummary(mlm_intslp_grp_pred_mtrx, params = params[!params %in% c("B")]) %>%
kableExtra::kable(
caption = "Raw Results: Varying-intercept & slope multilevel model, floor of measurement predictor + county-level (grp) uranium"
, digits = 4
) %>%
kableExtra::kable_styling(font_size = 12)| mean | sd | 2.5% | 50% | 97.5% | Rhat | n.eff | |
|---|---|---|---|---|---|---|---|
| G[1,1] | 1.4701 | 0.0389 | 1.3948 | 1.4698 | 1.5471 | 1.00 | 5757 |
| G[2,1] | -0.6887 | 0.0776 | -0.8410 | -0.6888 | -0.5351 | 1.00 | 3311 |
| G[1,2] | 0.7964 | 0.0972 | 0.6048 | 0.7984 | 0.9829 | 1.01 | 1380 |
| G[2,2] | -0.4091 | 0.2066 | -0.8199 | -0.4053 | -0.0343 | 1.04 | 277 |
| sigma.y | 0.7542 | 0.0187 | 0.7191 | 0.7538 | 0.7920 | 1.00 | 5328 |
| sigma.B[1] | 0.1588 | 0.0422 | 0.0852 | 0.1552 | 0.2508 | 1.01 | 442 |
| sigma.B[2] | 0.1616 | 0.1062 | 0.0244 | 0.1366 | 0.4130 | 1.12 | 127 |
| rho.B[1,1] | 1.0000 | 0.0000 | 1.0000 | 1.0000 | 1.0000 | NaN | 0 |
| rho.B[2,1] | 0.2304 | 0.3919 | -0.5795 | 0.2726 | 0.8420 | 1.01 | 386 |
| rho.B[1,2] | 0.2304 | 0.3919 | -0.5795 | 0.2726 | 0.8420 | 1.01 | 386 |
| rho.B[2,2] | 1.0000 | 0.0000 | 1.0000 | 1.0000 | 1.0000 | NaN | 0 |
Now clean results from matrix format
# function to name rows of G matrix
fn_g_mtrx_nm <- function(x_vars_nms, grp_vars_nms) {
tidyr::expand_grid(grp_vars_nms, x_vars_nms) %>%
mutate(
parameter = paste0("beta.", x_vars_nms, "_gamma.", grp_vars_nms)
) %>%
dplyr::select(parameter) %>%
c()
}
# function to name rows of sigma.B matrix
fn_sigmaB_mtrx_nm <- function(x_vars_nms) {
paste0("sigma.", x_vars_nms)
}
# nice table of results
dplyr::bind_rows(
# gamma matrix
MCMCvis::MCMCsummary(mlm_intslp_grp_pred_mtrx, params = params[params %in% c("G")]) %>%
as.data.frame() %>%
cbind(parameter = fn_g_mtrx_nm(x_vars_nms, grp_vars_nms)) %>%
dplyr::relocate(parameter) %>%
dplyr::arrange(parameter)
# sigma.B matrix
, MCMCvis::MCMCsummary(mlm_intslp_grp_pred_mtrx, params = params[params %in% c("sigma.B")]) %>%
as.data.frame() %>%
cbind(parameter = fn_sigmaB_mtrx_nm(x_vars_nms)) %>%
dplyr::relocate(parameter)
# sigma.y
, MCMCvis::MCMCsummary(mlm_intslp_grp_pred_mtrx, params = params[params %in% c("sigma.y", "rho.B")]) %>%
as.data.frame() %>%
tibble::rownames_to_column(var = "parameter")
) %>%
kableExtra::kable(
caption = "Cleaned Results: Varying-intercept & slope multilevel model, floor of measurement predictor + county-level (grp) uranium"
, digits = 4
, row.names = FALSE
) %>%
kableExtra::kable_styling(font_size = 12)| parameter | mean | sd | 2.5% | 50% | 97.5% | Rhat | n.eff |
|---|---|---|---|---|---|---|---|
| beta.intercept_gamma.intercept | 1.4701 | 0.0389 | 1.3948 | 1.4698 | 1.5471 | 1.00 | 5757 |
| beta.intercept_gamma.u | 0.7964 | 0.0972 | 0.6048 | 0.7984 | 0.9829 | 1.01 | 1380 |
| beta.x_gamma.intercept | -0.6887 | 0.0776 | -0.8410 | -0.6888 | -0.5351 | 1.00 | 3311 |
| beta.x_gamma.u | -0.4091 | 0.2066 | -0.8199 | -0.4053 | -0.0343 | 1.04 | 277 |
| sigma.intercept | 0.1588 | 0.0422 | 0.0852 | 0.1552 | 0.2508 | 1.01 | 442 |
| sigma.x | 0.1616 | 0.1062 | 0.0244 | 0.1366 | 0.4130 | 1.12 | 127 |
| sigma.y | 0.7542 | 0.0187 | 0.7191 | 0.7538 | 0.7920 | 1.00 | 5328 |
| rho.B[1,1] | 1.0000 | 0.0000 | 1.0000 | 1.0000 | 1.0000 | NaN | 0 |
| rho.B[2,1] | 0.2304 | 0.3919 | -0.5795 | 0.2726 | 0.8420 | 1.01 | 386 |
| rho.B[1,2] | 0.2304 | 0.3919 | -0.5795 | 0.2726 | 0.8420 | 1.01 | 386 |
| rho.B[2,2] | 1.0000 | 0.0000 | 1.0000 | 1.0000 | 1.0000 | NaN | 0 |
4.6 Non-nested models (p.289)
4.6.1 Example 1:
a psychological experiment with two potentially interacting factors
Data is from a psychological experiment of pilots on flight simulators, with \(n = 40\) data points corresponding to \(J = 5\) treatment conditions and \(K = 8\) different airports (e.g. sites, species, etc.). The responses can be fit to a non-nested multilevel model of the form:
\[\begin{align*} y_i &\sim \textrm{N}(\mu + \gamma_{j[i]} + \delta_{k[i]}, \sigma^2_y) \\ &\textrm{for} \; i = 1, \cdots, n \\ \gamma_j &\sim \textrm{N}(0, \sigma^2_{\gamma}) \\ &\textrm{for} \; j = 1, \cdots, J \\ \delta_k &\sim \textrm{N}(0, \sigma^2_{\delta}) \\ &\textrm{for} \; k = 1, \cdots, K \end{align*}\]
The parameters \(\gamma_j\) and \(\delta_k\) represent treatment effects and airport effects. Their distributions are centered at zero (rather than given mean levels \(\mu_\gamma\) and \(\mu_\delta\)) because the regression model for \(y\) already has an intercept, \(\mu\), and any nonzero mean for the \(\gamma\) and \(\delta\) distributions could be folded into \(\mu\). As we shall see in Section 19.4, it can sometimes be effective for computational purposes to add extra mean-level parameters into the model, but the coefficients in this expanded model must be interpreted with care. We can perform a quick fit as follows:
4.6.1.1 Load Data
# pilots <- read.table("http://www.stat.columbia.edu/~gelman/arm/examples/pilots/pilots.dat", header=TRUE, sep=",") %>%
pilots <- read.table("../data/pilots/pilots.dat", header=TRUE) %>%
dplyr::filter(recovered %in% c(0,1)) %>%
dplyr::group_by(group, scenario) %>%
dplyr::summarize(
successes = sum(recovered, na.rm = TRUE)
, trials = n()
) %>%
dplyr::ungroup() %>%
dplyr::mutate(
y = successes / trials
, treatment_index = as.numeric(as.factor(group))
, site_index = as.numeric(as.factor(scenario))
)
# scenario.abbr <- c("Nagoya", "B'ham", "Detroit", "Ptsbgh", "Roseln", "Chrlt", "Shemya", "Toledo")4.6.1.2 Using lme4::lmer (p. 289)
## Model fit
M1 <- lme4::lmer(data = pilots, formula = y ~ 1 + (1 | treatment_index) + (1 | site_index))
summary(M1)## Linear mixed model fit by REML ['lmerMod']
## Formula: y ~ 1 + (1 | treatment_index) + (1 | site_index)
## Data: pilots
##
## REML criterion at convergence: 12.3
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -1.30647 -0.66461 -0.07957 0.33792 2.96201
##
## Random effects:
## Groups Name Variance Std.Dev.
## site_index (Intercept) 0.10333 0.3214
## treatment_index (Intercept) 0.00000 0.0000
## Residual 0.04674 0.2162
## Number of obs: 40, groups: site_index, 8; treatment_index, 5
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 0.4418 0.1187 3.723
## optimizer (nloptwrap) convergence code: 0 (OK)
## boundary (singular) fit: see help('isSingular')4.6.1.3 Bayesian
4.6.1.3.1 Data and Initial conditions
## data for jags
data <- list(
n = nrow(pilots)
, J = pilots$treatment_index %>% unique() %>% length()
, K = pilots$site_index %>% unique() %>% length()
, y = pilots$y %>% as.double()
, treatment = pilots$treatment_index %>% as.double()
, site = pilots$site_index %>% as.double()
)
## inits for jags
inits <- function(){
list(
mu = rnorm(1, mean = 0, sd = 1)
, gamma = rnorm(n = data$J, mean = 0, sd = 1) # treatment effects (j)
, delta = rnorm(n = data$K, mean = 0, sd = 1) # site effects (k)
, sigma.gamma = runif(n = 1, min = 0, max = 1)
, sigma.delta = runif(n = 1, min = 0, max = 1)
, sigma.y = runif(n = 1, min = 0, max = 1)
)
}
# define parameters to return from MCMC
params <- c (
"mu"
, "gamma"
, "delta"
, "sigma.gamma"
, "sigma.delta"
, "sigma.y"
)4.6.1.4 JAGS Model
Recall model defined above
Write out the JAGS code for the model.
## JAGS Model
model {
###########################
# priors
###########################
# mu priors
mu ~ dnorm(0,1E-6)
# gamma priors
sigma.gamma ~ dunif(0, 100)
tau.gamma <- 1/sigma.gamma^2
mu.gamma <- 0
# delta priors
sigma.delta ~ dunif(0, 100)
tau.delta <- 1/sigma.delta^2
mu.delta <- 0
# y priors
sigma.y ~ dunif(0, 100)
tau.y <- 1/sigma.y^2
###########################
# likelihood
###########################
# treatment effects (j)
for (j in 1:J){
gamma[j] ~ dnorm(mu.gamma, tau.gamma)
}
# site effects (k)
for (k in 1:K){
delta[k] ~ dnorm(mu.delta, tau.delta)
}
# y
for (i in 1:n){
mu.y[i] <- mu + gamma[treatment[i]] + delta[site[i]]
y[i] ~ dnorm(mu.y[i], tau.y)
}
}4.6.1.5 Implement JAGS Model
##################################################################
# insert JAGS model code into an R script
##################################################################
{ # Extra bracket needed only for R markdown files - see answers
sink("pilots_pred1.R") # This is the file name for the jags code
cat("
## JAGS Model
model {
###########################
# priors
###########################
# mu priors
mu ~ dnorm(0,1E-6)
# gamma priors
sigma.gamma ~ dunif(0, 100)
tau.gamma <- 1/sigma.gamma^2
mu.gamma <- 0
# delta priors
sigma.delta ~ dunif(0, 100)
tau.delta <- 1/sigma.delta^2
mu.delta <- 0
# y priors
sigma.y ~ dunif(0, 100)
tau.y <- 1/sigma.y^2
###########################
# likelihood
###########################
# treatment effects (j)
for (j in 1:J){
gamma[j] ~ dnorm(mu.gamma, tau.gamma)
}
# site effects (k)
for (k in 1:K){
delta[k] ~ dnorm(mu.delta, tau.delta)
}
# y
for (i in 1:n){
mu.y[i] <- mu + gamma[treatment[i]] + delta[site[i]]
y[i] ~ dnorm(mu.y[i], tau.y)
}
}
", fill = TRUE)
sink()
}
##################################################################
# implement model
##################################################################
######################
# Call to JAGS
######################
pilots_pred1 = rjags::jags.model(
file = "pilots_pred1.R"
, data = data
, inits = inits
, n.chains = 3
, n.adapt = 100
)## Compiling model graph
## Resolving undeclared variables
## Allocating nodes
## Graph information:
## Observed stochastic nodes: 40
## Unobserved stochastic nodes: 17
## Total graph size: 191
##
## Initializing modelstats::update(pilots_pred1, n.iter = 1000, progress.bar = "none")
# save the coda object (more precisely, an mcmc.list object) to R
mlm_pilots_pred1 = rjags::coda.samples(
model = pilots_pred1
, variable.names = params
, n.iter = 1000
, n.thin = 1
, progress.bar = "none"
)4.6.1.6 Summary of the marginal posterior distributions of the parameters
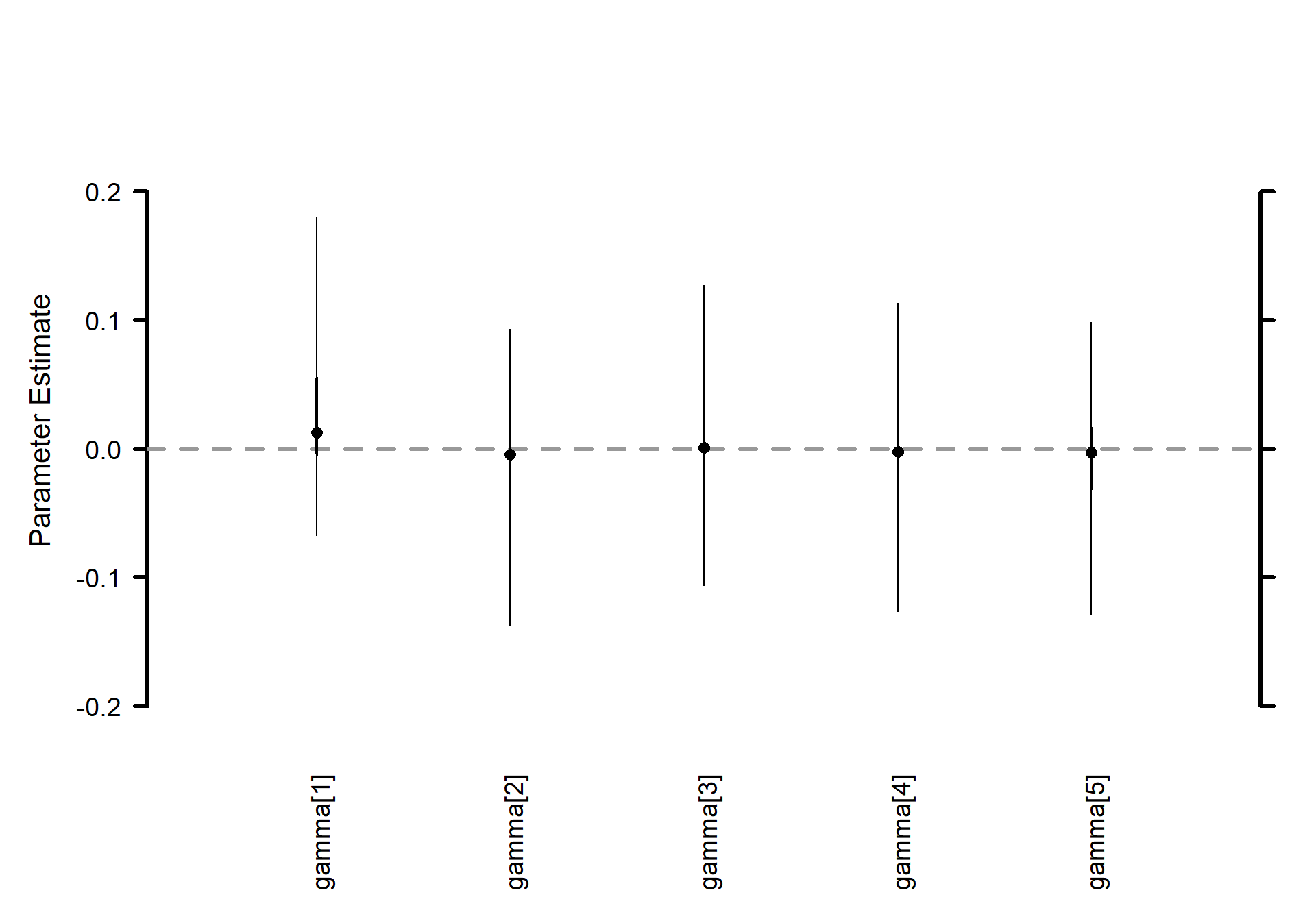
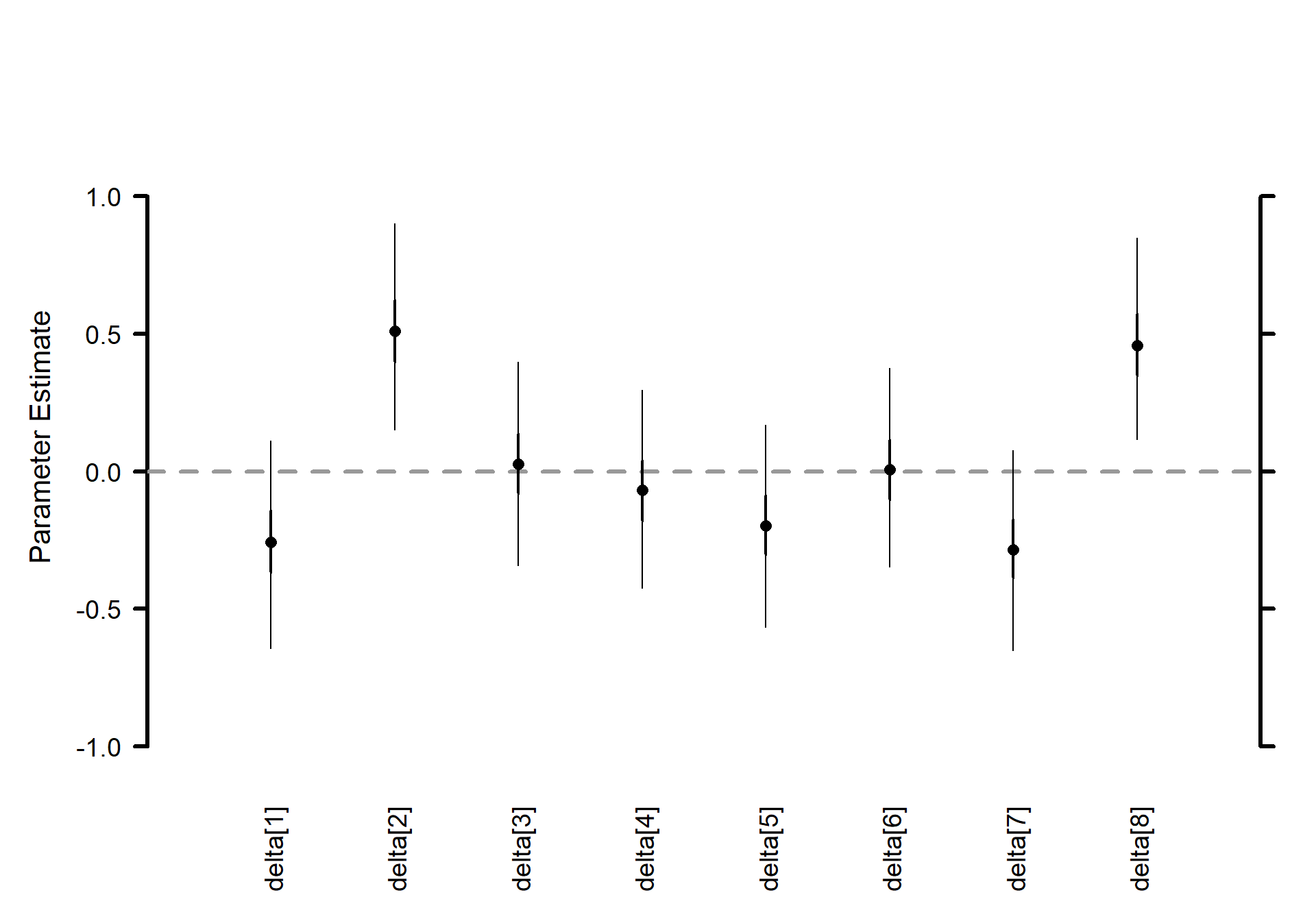
Thus, the variation among sites (airports) is huge \(\sigma_\delta\) – even larger than that among individual measurements \(\sigma_y\) – but the treatments \(\sigma_\gamma\) vary almost not at all.
# summary
MCMCvis::MCMCsummary(mlm_pilots_pred1, params = params[!params %in% c("gamma", "delta")]) %>%
kableExtra::kable(
caption = "Bayesian: treatment and site effects model"
, digits = 4
) %>%
kableExtra::kable_styling(font_size = 12)| mean | sd | 2.5% | 50% | 97.5% | Rhat | n.eff | |
|---|---|---|---|---|---|---|---|
| mu | 0.4126 | 0.1648 | 0.0851 | 0.4179 | 0.7381 | 1.05 | 90 |
| sigma.gamma | 0.0618 | 0.0518 | 0.0029 | 0.0483 | 0.1982 | 1.03 | 200 |
| sigma.delta | 0.4052 | 0.1726 | 0.2090 | 0.3693 | 0.8122 | 1.06 | 375 |
| sigma.y | 0.2270 | 0.0294 | 0.1776 | 0.2250 | 0.2911 | 1.00 | 1230 |
Treatment effects \(\boldsymbol{\gamma}\) (J=5) horizontal caterpillar plot
# Caterpillar plots
MCMCvis::MCMCplot(
mlm_pilots_pred1
, params = c("gamma")
, horiz = FALSE
, sz_med = 1.2, sz_thick = 2, sz_thin = 1
)
Site (airport) effects \(\boldsymbol{\delta}\) (K=8) horizontal caterpillar plot
# Caterpillar plots
MCMCvis::MCMCplot(
mlm_pilots_pred1
, params = c("delta")
, horiz = FALSE
, sz_med = 1.2, sz_thick = 2, sz_thin = 1
)
4.6.2 Example 2:
regression of earnings on ethnicity categories, age categories, and height
All the ideas of the earlier part of this chapter, introduced in the context of a simple structure of individuals within groups, apply to non-nested models as well. Here, we estimate a regression of log earnings, \(y_i\), on height, \(z_i\) (mean-adjusted), applied to the J = 4 ethnic groups (e.g. treatments) and K = 3 age categories (e.g. species). In essence, there is a separate regression model for each age group and ethnicity combination. The multilevel model can be written, somewhat awkwardly, as a data-level model,
\[ \begin{align*} y_i &\sim \textrm{N}(\alpha_{j[i],k[i]} + \beta_{j[i],k[i]} \cdot z_i , \sigma^2_y) \\ &\textrm{for} \; i = 1, \cdots, n \end{align*} \] a decomposition of the intercepts and slopes into terms for ethnicity, age, and ethnicity \(\times\) age,
\[ \begin{align*} \left(\begin{array}{c} \alpha_{j,k}\\ \beta_{j,k} \end{array}\right) &= \left(\begin{array}{c} \mu_{0}\\ \mu_{1} \end{array}\right) + \left(\begin{array}{c} \gamma_{0j}^\textrm{eth}\\ \gamma_{1j}^\textrm{eth} \end{array}\right) + \left(\begin{array}{c} \gamma_{0k}^\textrm{age}\\ \gamma_{1k}^\textrm{age} \end{array}\right) + \left(\begin{array}{c} \gamma_{0jk}^{\textrm{eth} \times \textrm{age}}\\ \gamma_{1jk}^{\textrm{eth} \times \textrm{age}} \end{array}\right) \end{align*} \]
and models for variation,
\[ \begin{align*} \left(\begin{array}{c} \gamma_{0j}^\textrm{eth}\\ \gamma_{1j}^\textrm{eth} \end{array}\right) &\sim \textrm{N}\left(\left(\begin{array}{c} 0\\ 0 \end{array}\right) , \Sigma^{\textrm{eth}} \right) \\ &\textrm{for} \; j = 1, \cdots, J \\ \left(\begin{array}{c} \gamma_{0k}^\textrm{age}\\ \gamma_{1k}^\textrm{age} \end{array}\right) &\sim \textrm{N}\left(\left(\begin{array}{c} 0\\ 0 \end{array}\right) , \Sigma^{\textrm{age}} \right) \\ &\textrm{for} \; k = 1, \cdots, K \\ \left(\begin{array}{c} \gamma_{0jk}^{\textrm{eth} \times \textrm{age}}\\ \gamma_{1jk}^{\textrm{eth} \times \textrm{age}} \end{array}\right) &\sim \textrm{N}\left(\left(\begin{array}{c} 0\\ 0 \end{array}\right) , \Sigma^{\textrm{eth} \times \textrm{age}} \right) \\ &\textrm{for} \; j = 1, \cdots, J \, \textrm{;} \, k = 1, \cdots, K \end{align*} \]
Because we have included means \(\mu_0\), \(\mu_1\) in the decomposition above, we can center each batch of coefficients at 0.
We fit this model, and the subsequent models in this chapter, in Bugs (see Chapter 17 for examples of code) because lme4::lmer() does not work so well when the number of groups is small—and, conversely, with these small datasets, Bugs is not too slow.